6ZZR
 
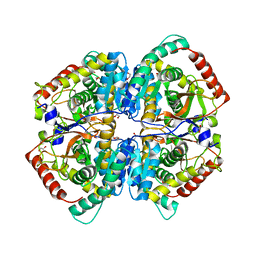 | | The Crystal Structure of human LDHA from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, L-lactate dehydrogenase A chain | | Authors: | Wang, F, Lin, D, Cheng, W, Bao, X, Zhu, B, Shang, H. | | Deposit date: | 2020-08-05 | | Release date: | 2020-08-19 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Crystal Structure of human LDHA from Biortus
To Be Published
|
|
8K46
 
 | | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses including all major Omicron strains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y, Gao, Y, Yao, H, Xu, W, Yang, H. | | Deposit date: | 2023-07-17 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses, including all major Omicron strains.
MedComm (2020), 4, 2023
|
|
8K45
 
 | | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses including all major Omicron strains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nb4 nanobody, ... | | Authors: | Lu, Y, Gao, Y, Yao, H, Xu, W, Yang, H. | | Deposit date: | 2023-07-17 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses, including all major Omicron strains.
MedComm (2020), 4, 2023
|
|
8K47
 
 | | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses including all major Omicron strains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y, Gao, Y, Yao, H, Xu, W, Yang, H. | | Deposit date: | 2023-07-17 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses, including all major Omicron strains.
MedComm (2020), 4, 2023
|
|
5E1L
 
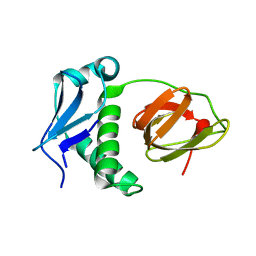 | | Structural and functional analysis of the E. coli FtsZ interacting protein, ZapC, reveals insight into molecular properties of a novel Z ring stabilizing protein | | Descriptor: | Cell division protein ZapC | | Authors: | Schumacher, M.A, Huang, K.-H, Tchorzewski, L, Zeng, W, Janakiraman, A. | | Deposit date: | 2015-09-29 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and Functional Analyses Reveal Insights into the Molecular Properties of the Escherichia coli Z Ring Stabilizing Protein, ZapC.
J.Biol.Chem., 291, 2016
|
|
7AAX
 
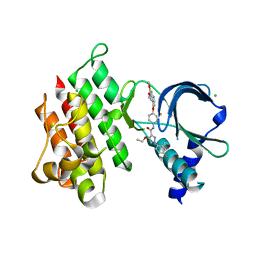 | | Crystal structure of MerTK kinase domain in complex with LDC1267 | | Descriptor: | CHLORIDE ION, Tyrosine-protein kinase Mer, ~{N}-[4-(6,7-dimethoxyquinolin-4-yl)oxy-3-fluoranyl-phenyl]-4-ethoxy-1-(4-fluoranyl-2-methyl-phenyl)pyrazole-3-carboxamide | | Authors: | Schimpl, M, Pflug, A, McCoull, W, Nissink, J.W.M, Overman, R.C, Rawlins, P.B, Truman, C, Underwood, E, Warwicker, J, Winter-Holt, J. | | Deposit date: | 2020-09-05 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.762 Å) | | Cite: | A-loop interactions in Mer tyrosine kinase give rise to inhibitors with two-step mechanism and long residence time of binding.
Biochem.J., 477, 2020
|
|
7AB0
 
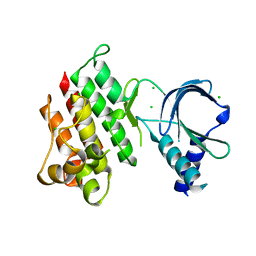 | | Apo crystal structure of the MerTK kinase domain | | Descriptor: | CHLORIDE ION, Tyrosine-protein kinase Mer | | Authors: | Pflug, A, Schimpl, M, McCoull, W, Nissink, J.W.M, Overman, R.C, Rawlins, P.B, Truman, C, Underwood, E, Warwicker, J, Winter-Holt, J. | | Deposit date: | 2020-09-05 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | A-loop interactions in Mer tyrosine kinase give rise to inhibitors with two-step mechanism and long residence time of binding.
Biochem.J., 477, 2020
|
|
4PM3
 
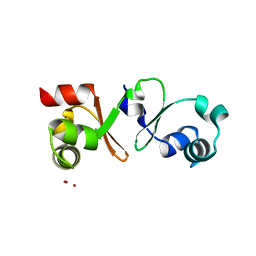 | |
8OHZ
 
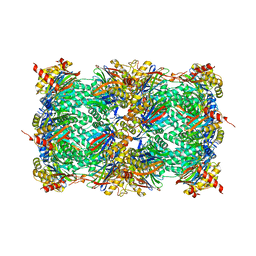 | | Yeast 20S proteasome in complex with a photoswitchable cepafungin derivative (transCep1) | | Descriptor: | (2~{S},3~{R})-2-[2-[4-[2-(4-ethylphenyl)hydrazinyl]phenyl]ethanoylamino]-~{N}-[(5~{S},8~{S},10~{S})-5-methyl-10-oxidanyl-2,7-bis(oxidanylidene)-1,6-diazacyclododec-8-yl]-3-oxidanyl-butanamide, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Morstein, J, Amatuni, A, Schuster, A, Kuttenlochner, W, Ko, T, Groll, M, Adibekian, A, Renata, H, Trauner, D.H. | | Deposit date: | 2023-03-21 | | Release date: | 2023-12-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Optical Control of Proteasomal Protein Degradation with a Photoswitchable Lipopeptide.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
5E4N
 
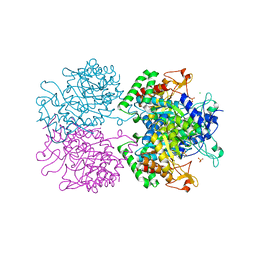 | | 3-Deoxy-D-arabino-heptulosonate 7-phosphate synthase from Mycobacterium tuberculosis with D-tyrosine bound in the tyrosine and phenylalanine binding sites | | Descriptor: | 3-deoxy-D-arabinoheptulosonate-7-phosphate synthase, CHLORIDE ION, D-TYROSINE, ... | | Authors: | Reichau, S, Jiao, W, Parker, E.J. | | Deposit date: | 2015-10-06 | | Release date: | 2016-06-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Probing the Sophisticated Synergistic Allosteric Regulation of Aromatic Amino Acid Biosynthesis in Mycobacterium tuberculosis Using -Amino Acids.
Plos One, 11, 2016
|
|
8OI1
 
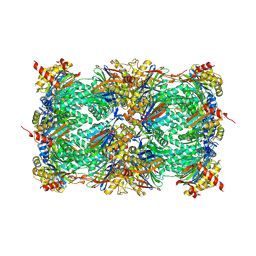 | | Yeast 20S proteasome in complex with a photoswitchable cepafungin derivative (transCep4) | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Morstein, J, Amatuni, A, Schuster, A, Kuttenlochner, W, Ko, T, Groll, M, Adibekian, A, Renata, H, Trauner, D.H. | | Deposit date: | 2023-03-21 | | Release date: | 2023-12-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Optical Control of Proteasomal Protein Degradation with a Photoswitchable Lipopeptide.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
6DTW
 
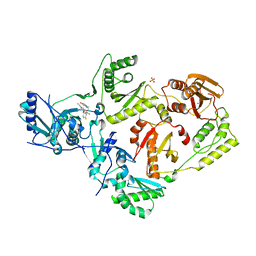 | | HIV-1 Reverse Transcriptase Y181C Mutant in complex with JLJ 578 | | Descriptor: | 8-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]-4-fluorophenoxy}-6-fluoroindolizine-2-carbonitrile, MAGNESIUM ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Sasaki, T, Gannam, Z.T.K, Anderson, K.S, Jorgensen, W.L, Lee, W. | | Deposit date: | 2018-06-18 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.742 Å) | | Cite: | Molecular and cellular studies evaluating a potent 2-cyanoindolizine catechol diether NNRTI targeting wildtype and Y181C mutant HIV-1 reverse transcriptase.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6U8U
 
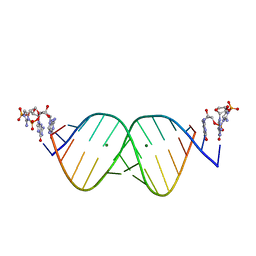 | | RNA duplex bound with TNA 3'-3' imidazolium dimer | | Descriptor: | 2-amino-3-[(R)-{[(3S,4R,5R)-5-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-4-hydroxyoxolan-3-yl]oxy}(hydroxy)phosphoryl]-1-[(S)-{[(3S,4R,5R)-5-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-4-hydroxyoxolan-3-yl]oxy}(hydroxy)phosphoryl]-1H-imidazol-3-ium, MAGNESIUM ION, RNA (5'-R(*(LCC)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*G)-3') | | Authors: | Szostak, J.W, Zhang, W. | | Deposit date: | 2019-09-05 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural interpretation of the effects of threo-nucleotides on nonenzymatic template-directed polymerization.
Nucleic Acids Res., 49, 2021
|
|
6UC0
 
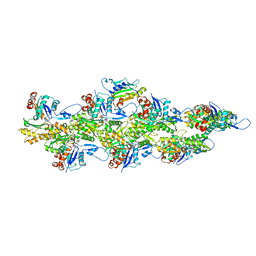 | | Isolated S3D-cofilin bound to an actin filament | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Huehn, A.R, Bibeau, J.P, Schramm, A.C, Cao, W, De La Cruz, E.M, Sindelar, C.V. | | Deposit date: | 2019-09-13 | | Release date: | 2020-01-01 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Structures of cofilin-induced structural changes reveal local and asymmetric perturbations of actin filaments.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5E2L
 
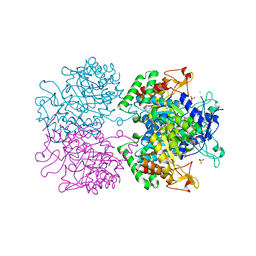 | | 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase from Mycobacterium tuberculosis in complex with D-phenylalanine | | Descriptor: | 3-deoxy-D-arabinoheptulosonate-7-phosphate synthase, CHLORIDE ION, D-PHENYLALANINE, ... | | Authors: | Reichau, S, Jiao, W, Parker, E.J. | | Deposit date: | 2015-10-01 | | Release date: | 2016-06-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Probing the Sophisticated Synergistic Allosteric Regulation of Aromatic Amino Acid Biosynthesis in Mycobacterium tuberculosis Using -Amino Acids.
Plos One, 11, 2016
|
|
2LZ3
 
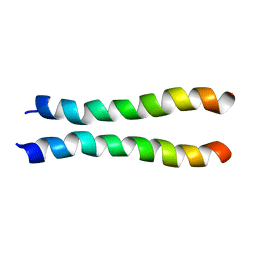 | |
6ZPL
 
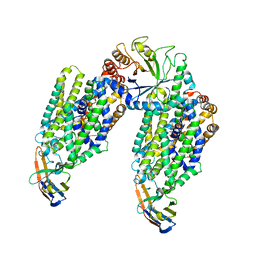 | | Inward-open structure of human glycine transporter 1 in complex with a benzoylisoindoline inhibitor, sybody Sb_GlyT1#7 and bound Na and Cl ions. | | Descriptor: | CHLORIDE ION, Endoglucanase H, SODIUM ION, ... | | Authors: | Shahsavar, A, Stohler, P, Bourenkov, G, Zimmermann, I, Siegrist, M, Guba, W, Pinard, E, Sinning, S, Seeger, M.A, Schneider, T.R, Dawson, R.J.P, Nissen, P. | | Deposit date: | 2020-07-08 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.945 Å) | | Cite: | Structural insights into the inhibition of glycine reuptake.
Nature, 591, 2021
|
|
8K5U
 
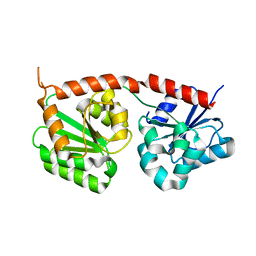 | | Se-glycosyltransferase RsSenB | | Descriptor: | Se glycosyltransferase | | Authors: | Huang, W, Long, F. | | Deposit date: | 2023-07-24 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of RsSenB at 2.15 Angstroms resolution
To Be Published
|
|
1QK1
 
 | | CRYSTAL STRUCTURE OF HUMAN UBIQUITOUS MITOCHONDRIAL CREATINE KINASE | | Descriptor: | CREATINE KINASE, UBIQUITOUS MITOCHONDRIAL, PHOSPHATE ION | | Authors: | Eder, M, Schlattner, U, Fritz-Wolf, K, Wallimann, T, Kabsch, W. | | Deposit date: | 1999-07-08 | | Release date: | 2000-04-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Human Ubiquitous Mitochondrial Creatine Kinase
Proteins: Struct.,Funct., Genet., 39, 2000
|
|
6UI7
 
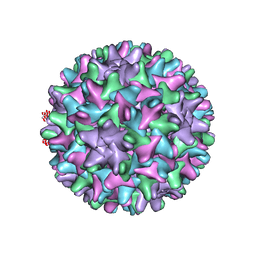 | | HBV T=4 149C3A | | Descriptor: | Core protein | | Authors: | Wu, W, Watts, N.R, Cheng, N, Huang, R, Steven, A, Wingfield, P.T. | | Deposit date: | 2019-09-30 | | Release date: | 2019-11-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Expression of quasi-equivalence and capsid dimorphism in the Hepadnaviridae.
Plos Comput.Biol., 16, 2020
|
|
2MJ2
 
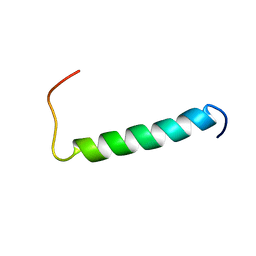 | | Structure of the dimerization domain of the human polyoma, JC virus agnoprotein is an amphipathic alpha-helix. | | Descriptor: | Agnoprotein | | Authors: | Coric, P, Saribas, S.A, Abou-Gharbia, M, Childers, W, White, M, Bouaziz, S, Safak, M. | | Deposit date: | 2013-12-23 | | Release date: | 2014-04-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structure of the dimerization domain of the human polyoma, JC virus agnoprotein is an amphipathic alpha-helix
J.Virol., 2014
|
|
1WCJ
 
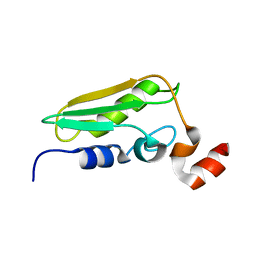 | | Conserved Hypothetical Protein TM0487 from Thermotoga maritima | | Descriptor: | HYPOTHETICAL PROTEIN TM0487 | | Authors: | Almeida, M.S, Peti, W, Herrmann, T, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2004-11-17 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Conserved Hypothetical Protein Tm0487 from Thermotoga Maritima: Implications for 216 Homologous Duf59 Proteins.
Protein Sci., 14, 2005
|
|
7A9L
 
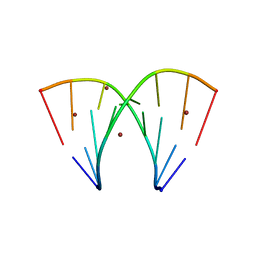 | | Racemic compound of RNA duplexes. | | Descriptor: | RNA (5'-R(*(0C)P*(0C)P*(0G)P*(0C)P*(0C)P*(0U)P*(0G)P*(0G))-3'), RNA (5'-R(*(0C)P*(0U)P*(0G)P*(0G)P*(0G)P*(0C)P*(0G)P*(0G))-3'), RNA (5'-R(*CP*CP*GP*CP*CP*UP*GP*G)-3'), ... | | Authors: | Rypniewski, W. | | Deposit date: | 2020-09-02 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Broken symmetry between RNA enantiomers in a crystal lattice.
Nucleic Acids Res., 49, 2021
|
|
7A9P
 
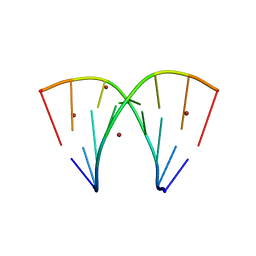 | | Racemic compound of RNA duplexes. | | Descriptor: | RNA (5'-R(*(0C)P*(0C)P*(0G)P*(0C)P*(0C)P*(0U)P*(0G)P*(0G))-3'), RNA (5'-R(*(0C)P*(0U)P*(0G)P*(0G)P*(0G)P*(0C)P*(0G)P*(0G))-3'), RNA (5'-R(*CP*CP*GP*CP*CP*UP*GP*G)-3'), ... | | Authors: | Rypniewski, W. | | Deposit date: | 2020-09-02 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.985 Å) | | Cite: | Broken symmetry between RNA enantiomers in a crystal lattice.
Nucleic Acids Res., 49, 2021
|
|
6ZWU
 
 | | Racemic compound of RNA duplexes. | | Descriptor: | RNA (5'-R(*(0C)P*(0C)P*(0G)P*(0C)P*(0C)P*(0U)P*(0G)P*(0G))-3'), RNA (5'-R(*(0C)P*(0U)P*(0G)P*(0G)P*(0G)P*(0C)P*(0G)P*(0G))-3'), RNA (5'-R(*CP*CP*GP*CP*CP*UP*GP*G)-3'), ... | | Authors: | Rypniewski, W. | | Deposit date: | 2020-07-28 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.527 Å) | | Cite: | Broken symmetry between RNA enantiomers in a crystal lattice.
Nucleic Acids Res., 49, 2021
|
|
