1U1F
 
 | | Structure of e. coli uridine phosphorylase complexed to 5-(m-(benzyloxy)benzyl)acyclouridine (BBAU) | | Descriptor: | 1-((2-HYDROXYETHOXY)METHYL)-5-(3-(BENZYLOXY)BENZYL)PYRIMIDINE-2,4(1H,3H)-DIONE, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Bu, W, Settembre, E.C, Ealick, S.E. | | Deposit date: | 2004-07-15 | | Release date: | 2005-07-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for inhibition of Escherichia coli uridine phosphorylase by 5-substituted acyclouridines.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1U2L
 
 | | Crystal structure of the C-terminal domain from the catalase-peroxidase KatG of Escherichia coli (P1) | | Descriptor: | Peroxidase/catalase HPI | | Authors: | Carpena, X, Melik-Adamyan, W, Loewen, P.C, Fita, I. | | Deposit date: | 2004-07-19 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the C-terminal domain of the catalase-peroxidase KatG from Escherichia coli.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2ACZ
 
 | | Complex II (Succinate Dehydrogenase) From E. Coli with Atpenin A5 inhibitor co-crystallized at the ubiquinone binding site | | Descriptor: | 3-[(2S,4S,5R)-5,6-DICHLORO-2,4-DIMETHYL-1-OXOHEXYL]-4-HYDROXY-5,6-DIMETHOXY-2(1H)-PYRIDINONE, CARDIOLIPIN, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Horsefield, R, Yankovskaya, V, Sexton, G, Whittingham, W, Shiomi, K, Omura, S, Byrne, B, Cecchini, G, Iwata, S. | | Deposit date: | 2005-07-19 | | Release date: | 2006-01-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and computational analysis of the quinone-binding site of complex II (succinate-ubiquinone oxidoreductase): a mechanism of electron transfer and proton conduction during ubiquinone reduction.
J.Biol.Chem., 281, 2006
|
|
1U3L
 
 | | IspF with Mg and CDP | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, CYTIDINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Steinbacher, S, Kaiser, J, Wungsintaweekul, J, Hechst, S, Eisenreich, W, Gerhardt, S, Bacher, A, Rohdich, F. | | Deposit date: | 2004-07-22 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of 2C-Methyl-D-Erythritol-2,4-Cyclodiphosphate Synthase Involved in Mevalonate Independent Biosynthesis of Isoprenoids
J.Mol.Biol., 316, 2002
|
|
2ADL
 
 | | Solution structure of the bacterial antitoxin CcdA: Implications for DNA and toxin binding | | Descriptor: | CcdA | | Authors: | Madl, T, VanMelderen, L, Oberer, M, Keller, W, Khatai, L, Zangger, K. | | Deposit date: | 2005-07-20 | | Release date: | 2006-08-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for nucleic acid and toxin recognition of the bacterial antitoxin CcdA
J.Mol.Biol., 364, 2006
|
|
2ADN
 
 | | Solution structure of the bacterial antitoxin CcdA: Implications for DNA and toxin binding | | Descriptor: | CcdA | | Authors: | Madl, T, VanMelderen, L, Oberer, M, Keller, W, Khatai, L, Zangger, K. | | Deposit date: | 2005-07-20 | | Release date: | 2006-08-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for nucleic acid and toxin recognition of the bacterial antitoxin CcdA
J.Mol.Biol., 364, 2006
|
|
2QTC
 
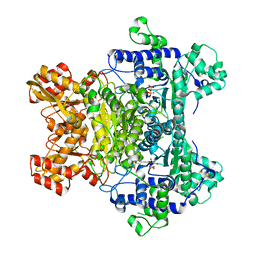 | | E. coli Pyruvate dehydrogenase E1 component E401K mutant with phosphonolactylthiamin diphosphate | | Descriptor: | 3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-{(1S)-1-HYDROXY-1-[(R)-HYDROXY(METHOXY)PHOSPHORYL]ETHYL}-5-(2-{[(S)-HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}ETHYL)-4-METHYL-1,3-THIAZOL-3-IUM, MAGNESIUM ION, Pyruvate dehydrogenase E1 component | | Authors: | Furey, W, Arjunan, P, Chandrasekhar, K. | | Deposit date: | 2007-08-01 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | A Dynamic Loop at the Active Center of the Escherichia coli Pyruvate Dehydrogenase Complex E1 Component Modulates Substrate Utilization and Chemical Communication with the E2 Component
J.Biol.Chem., 282, 2007
|
|
2ABY
 
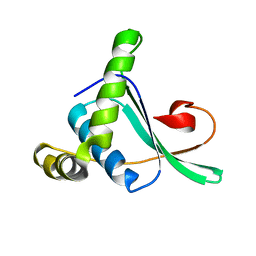 | | Solution structure of TA0743 from Thermoplasma acidophilum | | Descriptor: | hypothetical protein TA0743 | | Authors: | Kim, B, Jung, J, Hong, E, Yee, A, Arrowsmith, C.H, Lee, W. | | Deposit date: | 2005-07-18 | | Release date: | 2006-08-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the conserved novel-fold protein TA0743 from Thermoplasma acidophilum.
Proteins, 62, 2006
|
|
1U71
 
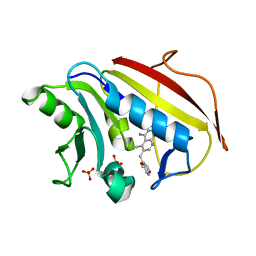 | | Understanding the Role of Leu22 Variants in Methotrexate Resistance: Comparison of Wild-type and Leu22Arg Variant Mouse and Human Dihydrofolate Reductase Ternary Crystal Complexes with Methotrexate and NADPH | | Descriptor: | 6-(2,5-DIMETHOXY-BENZYL)-5-METHYL-PYRIDO[2,3-D]PYRIMIDINE-2,4-DIAMINE, Dihydrofolate reductase, SULFATE ION | | Authors: | Cody, V, Luft, J.R, Pangborn, W. | | Deposit date: | 2004-08-02 | | Release date: | 2005-02-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Understanding the role of Leu22 variants in methotrexate resistance: comparison of wild-type and Leu22Arg variant mouse and human dihydrofolate reductase ternary crystal complexes with methotrexate and NADPH.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
3PEH
 
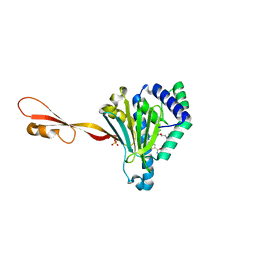 | | Crystal Structure of the N-terminal domain of an HSP90 from Plasmodium Falciparum, PFL1070c in the presence of a thienopyrimidine derivative | | Descriptor: | 2-amino-4-{2,4-dichloro-5-[2-(diethylamino)ethoxy]phenyl}-N-ethylthieno[2,3-d]pyrimidine-6-carboxamide, Endoplasmin homolog, SULFATE ION | | Authors: | Wernimont, A.K, Tempel, W, Hutchinson, A, Weadge, J, Cossar, D, MacKenzie, F, Vedadi, M, Senisterra, G, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Wyatt, P.G, Fairlamb, A.H, MacKenzie, C, Ferguson, M.A.J, Hui, R, Pizarro, J.C, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-10-26 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of the N-terminal domain of an HSP90 from Plasmodium Falciparum, PFL1070c in the presence of a thienopyrimidine derivative
To be Published
|
|
2QXG
 
 | | Crystal Structure of Human Kallikrein 7 in Complex with Ala-Ala-Phe-chloromethylketone | | Descriptor: | Kallikrein-7, L-alanyl-N-[(1S,2R)-1-benzyl-2-hydroxypropyl]-L-alaninamide | | Authors: | Debela, M, Hess, P, Magdolen, V, Bode, W, Steiner, T, Goettig, P. | | Deposit date: | 2007-08-11 | | Release date: | 2008-01-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Chymotryptic specificity determinants in the 1.0 A structure of the zinc-inhibited human tissue kallikrein 7.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2R0R
 
 | |
2AFP
 
 | | THE SOLUTION STRUCTURE OF TYPE II ANTIFREEZE PROTEIN REVEALS A NEW MEMBER OF THE LECTIN FAMILY | | Descriptor: | PROTEIN (SEA RAVEN TYPE II ANTIFREEZE PROTEIN) | | Authors: | Gronwald, W, Loewen, M.C, Lix, B, Daugulis, A.J, Sonnichsen, F.D, Davies, P.L, Sykes, B.D. | | Deposit date: | 1998-12-14 | | Release date: | 1998-12-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of type II antifreeze protein reveals a new member of the lectin family.
Biochemistry, 37, 1998
|
|
1VEY
 
 | | Crystal Structure of Peptide Deformylase from Leptospira Interrogans (LiPDF) at pH7.0 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Peptide deformylase, ZINC ION | | Authors: | Zhou, Z, Song, X, Li, Y, Gong, W. | | Deposit date: | 2004-04-06 | | Release date: | 2005-08-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Novel conformational states of peptide deformylase from pathogenic bacterium Leptospira interrogans: implications for population shift
J.Biol.Chem., 280, 2005
|
|
2A0X
 
 | | Structure of human purine nucleoside phosphorylase H257F mutant | | Descriptor: | 7-[[(3R,4R)-3-(hydroxymethyl)-4-oxidanyl-pyrrolidin-1-ium-1-yl]methyl]-3,5-dihydropyrrolo[3,2-d]pyrimidin-4-one, Purine nucleoside phosphorylase, SULFATE ION | | Authors: | Murkin, A.S, Shi, W, Schramm, V.L. | | Deposit date: | 2005-06-17 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Neighboring group participation in the transition state of human purine nucleoside phosphorylase.
Biochemistry, 46, 2007
|
|
2QDL
 
 | |
2A9J
 
 | |
2QMO
 
 | | Crystal structure of dethiobiotin synthetase (bioD) from Helicobacter pylori | | Descriptor: | CHLORIDE ION, Dethiobiotin synthetase | | Authors: | Chruszcz, M, Xu, X, Cuff, M, Cymborowski, M, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-16 | | Release date: | 2007-07-31 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural characterization of Helicobacter pylori dethiobiotin synthetase reveals differences between family members.
Febs J., 279, 2012
|
|
3PX0
 
 | |
1T6X
 
 | | Crystal structure of ADP bound TM379 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, riboflavin kinase/FMN adenylyltransferase | | Authors: | Shin, D.H, Wang, W, Kim, R, Yokota, H, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-05-07 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of ADP bound FAD synthetase
To be Published
|
|
2MTB
 
 | | Solution structure of apo_FldB | | Descriptor: | Flavodoxin-2 | | Authors: | Jin, C, Fu, W, Ye, Q. | | Deposit date: | 2014-08-15 | | Release date: | 2016-03-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Long-chain flavodoxin FldB from Escherichia coli
J.Biomol.Nmr, 60, 2014
|
|
3PFL
 
 | | CRYSTAL STRUCTURE OF PFL FROM E.COLI IN COMPLEX WITH SUBSTRATE ANALOGUE OXAMATE | | Descriptor: | OXAMIC ACID, PROTEIN (FORMATE ACETYLTRANSFERASE 1) | | Authors: | Becker, A, Fritz-Wolf, K, Kabsch, W, Knappe, J, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1999-05-14 | | Release date: | 2000-05-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and mechanism of the glycyl radical enzyme pyruvate formate-lyase.
Nat.Struct.Biol., 6, 1999
|
|
1T1U
 
 | | Structural Insights and Functional Implications of Choline Acetyltransferase | | Descriptor: | Choline O-acetyltransferase | | Authors: | Govindasamy, L, Pedersen, B, Lian, W, Kukar, T, Gu, Y, Jin, S, Agbandje-McKenna, M, Wu, D. | | Deposit date: | 2004-04-18 | | Release date: | 2005-04-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insights and functional implications of choline acetyltransferase
J.Struct.Biol., 148, 2004
|
|
2AJB
 
 | | Porcine dipeptidyl peptidase IV (CD26) in complex with the tripeptide tert-butyl-Gly-L-Pro-L-Ile (tBu-GPI) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-methyl-L-valyl-L-prolyl-L-isoleucine, ... | | Authors: | Engel, M, Hoffmann, T, Manhart, S, Heiser, U, Chambre, S, Huber, R, Demuth, H.U, Bode, W. | | Deposit date: | 2005-08-01 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Rigidity and flexibility of dipeptidyl peptidase IV: crystal structures of and docking experiments with DPIV.
J.Mol.Biol., 355, 2006
|
|
2HXT
 
 | | Crystal structure of L-Fuconate Dehydratase from Xanthomonas campestris liganded with Mg++ and D-erythronohydroxamate | | Descriptor: | (2R,3R)-N,2,3,4-TETRAHYDROXYBUTANAMIDE, L-fuconate dehydratase, MAGNESIUM ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Rakus, J.F, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2006-08-03 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evolution of Enzymatic Activities in the Enolase Superfamily: l-Fuconate Dehydratase from Xanthomonas campestris.
Biochemistry, 45, 2006
|
|
