3K5O
 
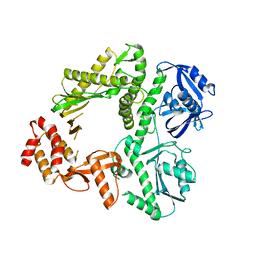 | | Crystal structure of E.coli Pol II | | Descriptor: | DNA polymerase II | | Authors: | Yang, W, Wang, F. | | Deposit date: | 2009-10-07 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insight into translesion synthesis by DNA Pol II.
Cell(Cambridge,Mass.), 139, 2009
|
|
1XDK
 
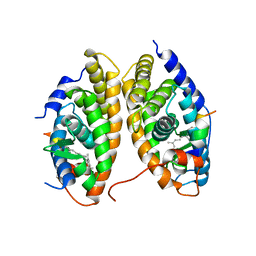 | | Crystal Structure of the RARbeta/RXRalpha Ligand Binding Domain Heterodimer in Complex with 9-cis Retinoic Acid and a Fragment of the TRAP220 Coactivator | | Descriptor: | (9cis)-retinoic acid, Retinoic acid receptor RXR-alpha, Retinoic acid receptor, ... | | Authors: | Pogenberg, V, Guichou, J.F, Vivat-Hannah, V, Kammerer, S, Perez, E, Germain, P, De Lera, A.R, Gronemeyer, H, Royer, C.A, Bourguet, W. | | Deposit date: | 2004-09-07 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | CHARACTERIZATION OF THE INTERACTION BETWEEN RAR/RXR HETERODIMERS AND TRANSCRIPTIONAL COACTIVATORS THROUGH STRUCTURAL AND FLUORESCENCE ANISOTROPY STUDIES
J.Biol.Chem., 280, 2005
|
|
7TUB
 
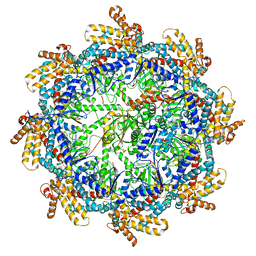 | | The beta-tubulin folding intermediate IV | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Zhao, Y, Frydman, J, Chiu, W. | | Deposit date: | 2022-02-02 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural visualization of the tubulin folding pathway directed by human chaperonin TRiC/CCT.
Cell, 185, 2022
|
|
7TRG
 
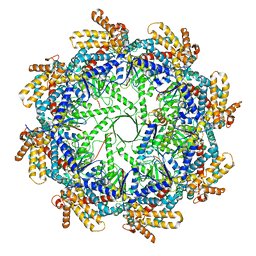 | | The beta-tubulin folding intermediate I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Zhao, Y, Frydman, J, Chiu, W. | | Deposit date: | 2022-01-28 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural visualization of the tubulin folding pathway directed by human chaperonin TRiC/CCT.
Cell, 185, 2022
|
|
7TTN
 
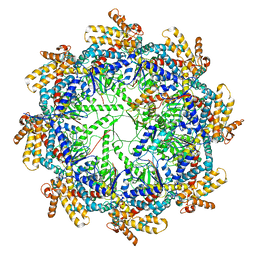 | | The beta-tubulin folding intermediate II | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Zhao, Y, Frydman, J, Chiu, W. | | Deposit date: | 2022-02-01 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural visualization of the tubulin folding pathway directed by human chaperonin TRiC/CCT.
Cell, 185, 2022
|
|
7TTT
 
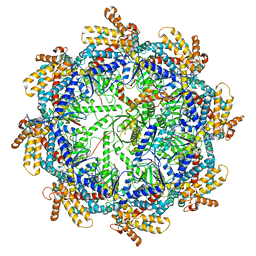 | | The beta-tubulin folding intermediate III | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Zhao, Y, Frydman, J, Chiu, W. | | Deposit date: | 2022-02-01 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural visualization of the tubulin folding pathway directed by human chaperonin TRiC/CCT.
Cell, 185, 2022
|
|
3JT1
 
 | | Legionella pneumophila glucosyltransferase Lgt1, UDP-bound form | | Descriptor: | Putative uncharacterized protein, URIDINE-5'-DIPHOSPHATE | | Authors: | Lu, W, Du, J, Belyi, Y, Stahl, M, Zivilikidis, T, Gerhardt, S, Aktories, K, Einsle, O. | | Deposit date: | 2009-09-11 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of the Action of Glucosyltransferase Lgt1 from Legionella pneumophila.
J.Mol.Biol., 2009
|
|
1XK8
 
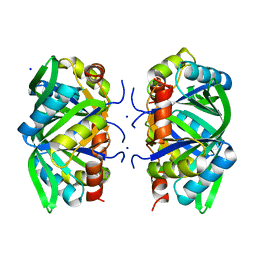 | | Divalent cation tolerant protein CUTA from Homo sapiens O60888 | | Descriptor: | Divalent cation tolerant protein CUTA, SODIUM ION | | Authors: | Tempel, W, Chen, L, Liu, Z.-J, Lee, D, Shah, A, Dailey, T.A, Mayer, M.R, Arendall III, W.B, Rose, J.P, Dailey, H.A, Richardson, J.S, Richardson, D.C, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-27 | | Release date: | 2004-10-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Divalent cation tolerant protein CUTA from Homo sapiens O60888
To be published
|
|
1XHC
 
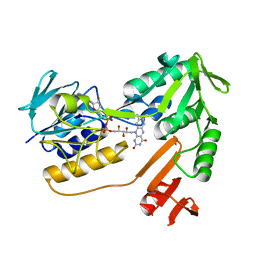 | | NADH oxidase /nitrite reductase from Pyrococcus furiosus Pfu-1140779-001 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH oxidase /nitrite reductase | | Authors: | Horanyi, P, Tempel, W, Weinberg, M.V, Liu, Z.-J, Shah, A, Chen, L, Lee, D, Sugar, F.J, Brereton, P.S, Izumi, M, Poole II, F.L, Shah, C, Jenney Jr, F.E, Arendall III, W.B, Rose, J.P, Adams, M.W.W, Richardson, J.S, Richardson, D.C, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-17 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | NADH oxidase /nitrite reductase from Pyrococcus furiosus Pfu-1140779-001
To be published
|
|
4PIP
 
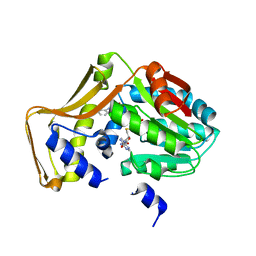 | | Engineered EgtD variant EgtD-M252V,E282A in complex with tryptophan and SAH | | Descriptor: | CHLORIDE ION, Histidine-specific methyltransferase EgtD, MAGNESIUM ION, ... | | Authors: | Vit, A, Seebeck, F.P, Blankenfeldt, W. | | Deposit date: | 2014-05-09 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ergothioneine Biosynthetic Methyltransferase EgtD Reveals the Structural Basis of Aromatic Amino Acid Betaine Biosynthesis.
Chembiochem, 16, 2015
|
|
3JAB
 
 | | Domain organization and conformational plasticity of the G protein effector, PDE6 | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, GafA domain of cone phosphodiesterase 6C, GafB domain of phosphodiesterase 2A, ... | | Authors: | Zhang, Z, He, F, Constantine, R, Baker, M.L, Baehr, W, Schmid, M.F, Wensel, T.G, Agosto, M.A. | | Deposit date: | 2015-05-26 | | Release date: | 2015-06-10 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Domain Organization and Conformational Plasticity of the G Protein Effector, PDE6.
J.Biol.Chem., 290, 2015
|
|
3JYN
 
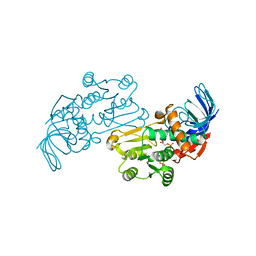 | | Crystal structures of Pseudomonas syringae pv. Tomato DC3000 quinone oxidoreductase complexed with NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Quinone oxidoreductase | | Authors: | Pan, X, Zhang, H, Gao, Y, Li, M, Chang, W. | | Deposit date: | 2009-09-22 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structures of Pseudomonas syringae pv. tomato DC3000 quinone oxidoreductase and its complex with NADPH
Biochem.Biophys.Res.Commun., 390, 2009
|
|
1X71
 
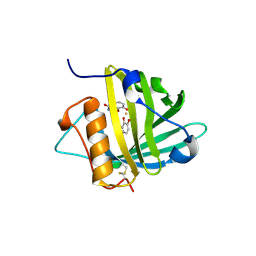 | | Crystal structure of Siderocalin (NGAL, Lipocalin 2) complexed with TRENCAM-3,2-HOPO, a cepabactin analogue | | Descriptor: | 2,3-DIHYDROXYBENZAMIDE, FE (III) ION, Neutrophil gelatinase-associated lipocalin | | Authors: | Holmes, M.A, Paulsene, W, Jide, X, Ratledge, C, Strong, R.K. | | Deposit date: | 2004-08-12 | | Release date: | 2005-01-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Siderocalin (Lcn 2) Also Binds Carboxymycobactins, Potentially Defending against Mycobacterial Infections through Iron Sequestration
Structure, 13, 2005
|
|
1X89
 
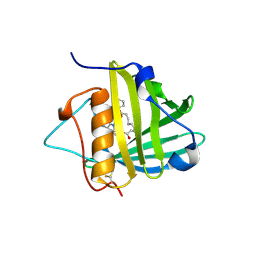 | | Crystal structure of Siderocalin (NGAL, Lipocalin 2) complexed with Carboxymycobactin S | | Descriptor: | CARBOXYMYCOBACTIN S, Neutrophil gelatinase-associated lipocalin | | Authors: | Holmes, M.A, Paulsene, W, Jide, X, Ratledge, C, Strong, R.K. | | Deposit date: | 2004-08-17 | | Release date: | 2005-01-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Siderocalin (Lcn 2) Also Binds Carboxymycobactins, Potentially Defending against Mycobacterial Infections through Iron Sequestration
Structure, 13, 2005
|
|
1XC1
 
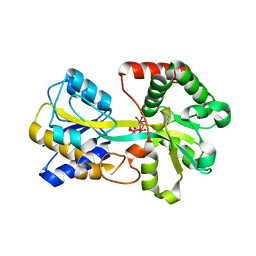 | | Oxo Zirconium(IV) Cluster in the Ferric Binding Protein (FBP) | | Descriptor: | OXO ZIRCONIUM(IV) CLUSTER, periplasmic iron-binding protein | | Authors: | Zhong, W, Alexeev, D, Harvey, I, Guo, M, Hunter, D.J.B, Zhu, H, Campopiano, D.J, Sadler, P.J. | | Deposit date: | 2004-08-31 | | Release date: | 2004-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Assembly of an Oxo-Zirconium(IV) Cluster in a Protein Cleft
Angew.Chem.Int.Ed.Engl., 43, 2004
|
|
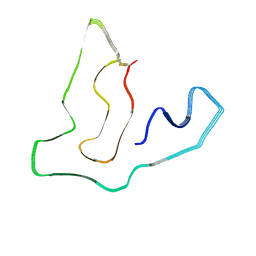
7TMC
 
 | |
3IST
 
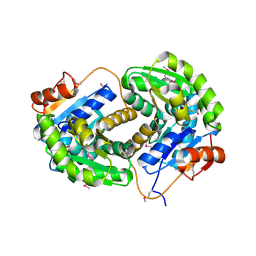 | | Crystal structure of glutamate racemase from Listeria monocytogenes in complex with succinic acid | | Descriptor: | CHLORIDE ION, Glutamate racemase, SUCCINIC ACID | | Authors: | Majorek, K.A, Chruszcz, M, Skarina, T, Onopriyenko, O, Stam, J, Anderson, W.F, Savchenko, A, Bujnicki, J.M, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-27 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of glutamate racemase from Listeria monocytogenes in complex with succinic acid
TO BE PUBLISHED
|
|
1XI6
 
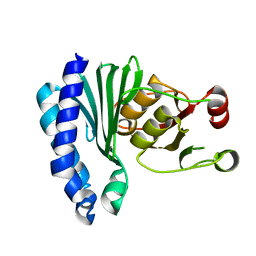 | | Extragenic suppressor from Pyrococcus furiosus Pfu-1862794-001 | | Descriptor: | extragenic suppressor | | Authors: | Zhao, M, Chang, J.C, Zhou, W, Chen, L, Horanyi, P, Xu, H, Yang, H, Liu, Z.-J, Habel, J.E, Lee, D, Chang, S.-H, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-21 | | Release date: | 2004-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Extragenic suppressor from Pyrococcus furiosus Pfu-1862794-001
To be published
|
|
4PIM
 
 | |
1XH6
 
 | | Crystal Structures of Protein Kinase B Selective Inhibitors in Complex with Protein Kinase A and Mutants | | Descriptor: | N-(4-{[4-(2-HYDROXY-5-PIPERIDIN-1-YLBENZOYL)BENZOYL]AMINO}AZEPAN-3-YL)ISONICOTINAMIDE, cAMP-dependent protein kinase inhibitor, alpha form, ... | | Authors: | Breitenlechner, C.B, Friebe, W.-G, Brunet, E, Werner, G, Graul, K, Thomas, U, Kuenkele, K.-P, Schaefer, W, Gassel, M, Bossemeyer, D, Huber, R, Engh, R.A, Masjost, B. | | Deposit date: | 2004-09-17 | | Release date: | 2005-09-17 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design and crystal structures of protein kinase B-selective inhibitors in complex with protein kinase A and mutants
J.Med.Chem., 48, 2005
|
|
3J02
 
 | | Lidless D386A Mm-cpn in the pre-hydrolysis ATP-bound state | | Descriptor: | Lidless D386A Mm-cpn variant | | Authors: | Zhang, J, Ma, B, DiMaio, F, Douglas, N.R, Joachimiak, L, Baker, D, Frydman, J, Levitt, M, Chiu, W. | | Deposit date: | 2011-02-10 | | Release date: | 2011-05-18 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Cryo-EM structure of a group II chaperonin in the prehydrolysis ATP-bound state leading to lid closure.
Structure, 19, 2011
|
|
2VU2
 
 | | Biosynthetic thiolase from Z. ramigera. Complex with S-pantetheine-11- pivalate. | | Descriptor: | (3R)-3-hydroxy-2,2-dimethyl-4-oxo-4-({3-oxo-3-[(2-sulfanylethyl)amino]propyl}amino)butyl 2,2-dimethylpropanoate, ACETYL-COA ACETYLTRANSFERASE, SULFATE ION | | Authors: | Kursula, P, Merilainen, G, Schmitz, W, Wierenga, R.K. | | Deposit date: | 2008-05-19 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Sulfur Atoms of the Substrate Coa and the Catalytic Cysteine are Required for a Productive Mode of Substrate Binding in Bacterial Biosynthetic Thiolase, a Thioester-Dependent Enzyme.
FEBS J., 275, 2008
|
|
2VXS
 
 | | Structure of IL-17A in complex with a potent, fully human neutralising antibody | | Descriptor: | FAB FRAGMENT, INTERLEUKIN-17A, SULFATE ION | | Authors: | Gerhardt, S, Hargreaves, D, Pauptit, R.A, Davies, R.A, Russell, C, Welsh, F, Tuske, S.J, Coales, S.J, Hamuro, Y, Needham, M.R.C, Langham, C, Barker, W, Bell, P, Aziz, A, Smith, M.J, Dawson, S, Abbott, W.M. | | Deposit date: | 2008-07-09 | | Release date: | 2009-07-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structure of Il-17A in Complex with a Potent, Fully Human Neutralising Antibody.
J.Mol.Biol., 394, 2009
|
|
1EXE
 
 | |
3J0B
 
 | | cryo-EM reconstruction of West Nile virus | | Descriptor: | envelope glycoprotein E | | Authors: | Zhang, W, Kaufmann, B, Chipman, P.R, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2011-06-15 | | Release date: | 2012-12-19 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (10.3 Å) | | Cite: | Membrane curvature in flaviviruses.
J.Struct.Biol., 183, 2013
|
|
