2WFD
 
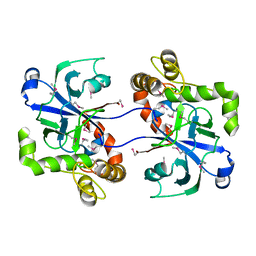 | | Structure of the human cytosolic leucyl-tRNA synthetase editing domain | | Descriptor: | LEUCYL-TRNA SYNTHETASE, CYTOPLASMIC | | Authors: | Seiradake, E, Mao, W, Hernandez, V, Baker, S.J, Plattner, J.J, Alley, M.R.K, Cusack, S. | | Deposit date: | 2009-04-03 | | Release date: | 2009-05-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal Structures of the Human and Fungal Cytosolic Leucyl-tRNA Synthetase Editing Domains: A Structural Basis for the Rational Design of Antifungal Benzoxaboroles.
J.Mol.Biol., 390, 2009
|
|
2C8Y
 
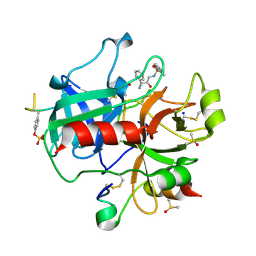 | | thrombin inhibitors | | Descriptor: | DIMETHYL SULFOXIDE, HIRUDIN VARIANT-2, N-[(2R,3S)-3-AMINO-2-HYDROXY-4-PHENYLBUTYL]NAPHTHALENE-2-SULFONAMIDE, ... | | Authors: | Howard, N, Abell, C, Blakemore, W, Carr, R, Chessari, G, Congreve, M, Howard, S, Jhoti, H, Murray, C.W, Seavers, L.C.A, van Montfort, R.L.M. | | Deposit date: | 2005-12-08 | | Release date: | 2006-07-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Application of Fragment Screening and Fragment Linking to the Discovery of Novel Thrombin Inhibitors
J.Med.Chem., 49, 2006
|
|
2EIG
 
 | | Lotus tetragonolobus seed lectin (Isoform) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, MANGANESE (II) ION, ... | | Authors: | Moreno, F.B.M.B, Vicoti, M.M, Abrego, J.R.B, de Oliveira, T.M, Bezerra, G.A, Cavada, B.S, Filgueira de Azevedo Jr, W. | | Deposit date: | 2007-03-13 | | Release date: | 2008-03-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of a new quaternary association for legume lectins
J.Struct.Biol., 161, 2008
|
|
2E6B
 
 | |
8PKI
 
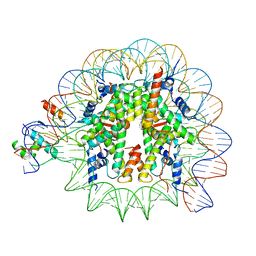 | | Cryo-EM structure of NR5A2-nucleosome complex SHL+5.5 | | Descriptor: | DNA, Histone H2A, Histone H2B type 1-C/E/G, ... | | Authors: | Kobayashi, W, Sappler, A, Bollschweiler, D, Kummecke, M, Basquin, J, Arslantas, E, Ruangroengkulrith, S, Hornberger, R, Duderstadt, K, Tachibana, K. | | Deposit date: | 2023-06-26 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Nucleosome-bound NR5A2 structure reveals pioneer factor mechanism by DNA minor groove anchor competition.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8PKJ
 
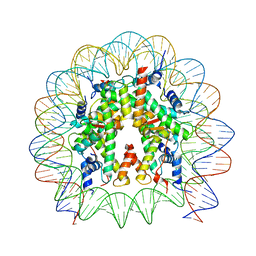 | | Cryo-EM structure of the nucleosome containing Nr5a2 motif at SHL+5.5 | | Descriptor: | DNA, Histone H2A, Histone H2B, ... | | Authors: | Kobayashi, W, Sappler, A, Bollschweiler, D, Kummecke, M, Basquin, J, Arslantas, E, Ruangroengkulrith, S, Hornberger, R, Duderstadt, K, Tachibana, K. | | Deposit date: | 2023-06-26 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Nucleosome-bound NR5A2 structure reveals pioneer factor mechanism by DNA minor groove anchor competition.
Nat.Struct.Mol.Biol., 31, 2024
|
|
2X4D
 
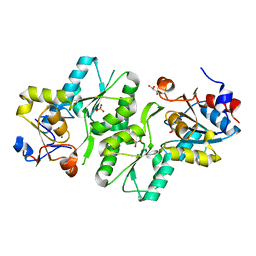 | | Crystal structure of human phospholysine phosphohistidine inorganic pyrophosphate phosphatase LHPP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Vollmar, M, Gileadi, C, Guo, K, Savitsky, P, Muniz, J.R.C, Yue, W, Allerston, C, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Kavanagh, K.L, Oppermann, U. | | Deposit date: | 2010-01-29 | | Release date: | 2010-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of Human Phospholysine Phosphohistidine Inorganic Pyrophosphate Phosphatase Lhpp
To be Published
|
|
3U2Z
 
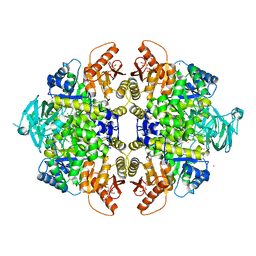 | | Activator-Bound Structure of Human Pyruvate Kinase M2 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 6-(3-aminobenzyl)-4-methyl-2-methylsulfinyl-4,6-dihydro-5H-thieno[2',3':4,5]pyrrolo[2,3-d]pyridazin-5-one, Pyruvate kinase isozymes M1/M2, ... | | Authors: | Hong, B, Dimov, S, Tempel, W, Auld, D, Thomas, C, Boxer, M, Jianq, J.-K, Skoumbourdis, A, Min, S, Southall, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Inglese, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-10-04 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis.
Nat.Chem.Biol., 8, 2012
|
|
2WJG
 
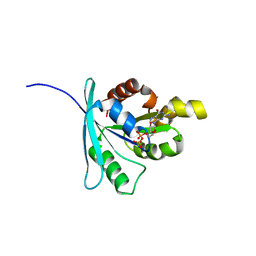 | | Structure and function of the FeoB G-domain from Methanococcus jannaschii | | Descriptor: | FERROUS IRON TRANSPORT PROTEIN B HOMOLOG, GUANOSINE-5'-DIPHOSPHATE, POLYALANINE | | Authors: | Koester, S, Wehner, M, Herrmann, C, Kuehlbrandt, W, Yildiz, O. | | Deposit date: | 2009-05-26 | | Release date: | 2009-07-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Function of the Feob G-Domain from Methanococcus Jannaschii.
J.Mol.Biol., 392, 2009
|
|
2E5O
 
 | | 'Solution structure of the TRIP_4C domain of target of activating signal cointegrator 1 | | Descriptor: | Activating signal cointegrator 1 | | Authors: | Dang, W, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-12-22 | | Release date: | 2008-01-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the TRIP_4C domain of target of activating signal cointegrator 1
To be Published
|
|
2E6E
 
 | |
2E69
 
 | |
8PJO
 
 | | Cryo EM structure of the type 3D polymorph of alpha-synuclein E46K mutant at low pH. | | Descriptor: | Alpha-synuclein, CHLORIDE ION | | Authors: | Frey, L, Qureshi, B.M, Kwiatkowski, W, Rhyner, D, Greenwald, J, Riek, R. | | Deposit date: | 2023-06-23 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.31 Å) | | Cite: | On the pH-dependence of alpha-synuclein amyloid polymorphism and the role of secondary nucleation in seed-based amyloid propagation
Elife, 2023
|
|
8PK4
 
 | | Cryo EM structure of the type 5A polymorph of alpha-synuclein. | | Descriptor: | Alpha-synuclein | | Authors: | Frey, L, Qureshi, B.M, Kwiatkowski, W, Rhyner, D, Greenwald, J, Riek, R. | | Deposit date: | 2023-06-24 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | On the pH-dependence of alpha-synuclein amyloid polymorphism and the role of secondary nucleation in seed-based amyloid propagation
Elife, 2023
|
|
3U3S
 
 | | The S-SAD phased crystal structure of the ecto-domain of Death Receptor 6 (DR6) | | Descriptor: | Tumor necrosis factor receptor superfamily member 21 | | Authors: | Ru, H, Zhao, L.X, Ding, W, Jiao, L.Y, Shaw, N, Zhang, L.G, Hung, L.W, Matsugaki, N, Wakatsuki, S, Liu, Z.J. | | Deposit date: | 2011-10-06 | | Release date: | 2012-05-02 | | Last modified: | 2013-07-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | S-SAD phasing study of death receptor 6 and its solution conformation revealed by SAXS.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1CFQ
 
 | | ANTI-P24 (HIV-1) FAB FRAGMENT CB41 | | Descriptor: | PROTEIN (IGG2A KAPPA ANTIBODY CB41 (HEAVY CHAIN)), PROTEIN (IGG2A KAPPA ANTIBODY CB41 (LIGHT CHAIN)) | | Authors: | Keitel, T, Kramer, A, Wessner, H, Scholz, C, Schneider-Mergener, J, Hoehne, W. | | Deposit date: | 1999-03-19 | | Release date: | 1999-03-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallographic analysis of anti-p24 (HIV-1) monoclonal antibody cross-reactivity and polyspecificity.
Cell(Cambridge,Mass.), 91, 1997
|
|
8PK2
 
 | | Cryo EM structure of the type 1m polymorph of alpha-synuclein | | Descriptor: | Alpha-synuclein | | Authors: | Frey, L, Qureshi, B.M, Kwiatkowski, W, Rhyner, D, Greenwald, J, Riek, R. | | Deposit date: | 2023-06-24 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | On the pH-dependence of alpha-synuclein amyloid polymorphism and the role of secondary nucleation in seed-based amyloid propagation
Elife, 2023
|
|
2WO7
 
 | | Isopenicillin N synthase with substrate analogue L,L,D-ACd2Ab (unexposed) | | Descriptor: | DELTA-(L-ALPHA-AMINOADIPOYL)-L-CYSTEINYL-D-VINYLGLYCINE, FE (II) ION, ISOPENICILLIN N SYNTHASE | | Authors: | Ge, W, Clifton, I.J, Adlington, R.M, Baldwin, J.E, Rutledge, P.J. | | Deposit date: | 2009-07-22 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic Studies on the Binding of Selectively Deuterated Lld- and Lll-Substrate Epimers by Isopenicillin N Synthase.
Biochem.Biophys.Res.Commun., 398, 2010
|
|
2E71
 
 | | Solution structure of the second FF domain of human transcription factor CA150 | | Descriptor: | Transcription elongation regulator 1 | | Authors: | Tanabe, W, Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-05 | | Release date: | 2007-07-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second FF domain of human transcription factor CA150
To be Published
|
|
2B8Y
 
 | | Crystal structure of the interleukin-4 variant T13DF82D | | Descriptor: | Interleukin-4, SULFATE ION | | Authors: | Kraich, M, Klein, M, Patino, E, Harrer, H, Sebald, W, Mueller, T.D. | | Deposit date: | 2005-10-10 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A modular interface of IL-4 allows for scalable affinity without affecting specificity for the IL-4 receptor
Bmc Biol., 4, 2006
|
|
8QQ0
 
 | | SARS-CoV-2 S protein bound to neutralising antibody UZGENT_A3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Remaut, H, Reiter, D, Vandenkerckhove, L, Acar, D.D, Witkowski, W, Gerlo, S. | | Deposit date: | 2023-10-03 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Integrating artificial intelligence-based epitope prediction in a SARS-CoV-2 antibody discovery pipeline: caution is warranted.
Ebiomedicine, 100, 2024
|
|
2B91
 
 | | Crystal structure of the interleukin-4 variant F82DR85A | | Descriptor: | Interleukin-4, SULFATE ION | | Authors: | Kraich, M, Klein, M, Patino, E, Harrer, H, Sebald, W, Mueller, T.D. | | Deposit date: | 2005-10-10 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A modular interface of IL-4 allows for scalable affinity without affecting specificity for the IL-4 receptor
Bmc Biol., 4, 2006
|
|
2B8X
 
 | | Crystal structure of the interleukin-4 variant F82D | | Descriptor: | Interleukin-4, SULFATE ION | | Authors: | Kraich, M, Klein, M, Patino, E, Harrer, H, Sebald, W, Mueller, T.D. | | Deposit date: | 2005-10-10 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A modular interface of IL-4 allows for scalable affinity without affecting specificity for the IL-4 receptor
Bmc Biol., 4, 2006
|
|
2BCO
 
 | | X-ray structure of succinylglutamate desuccinalase from Vibrio Parahaemolyticus (RIMD 2210633) at the resolution 2.3 A, Northeast Structural Genomics Target Vpr14 | | Descriptor: | Succinylglutamate desuccinylase, ZINC ION | | Authors: | Kuzin, A.P, Abashidze, M, Forouhar, F, Benach, J, Zhou, W, Acton, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-10-19 | | Release date: | 2005-10-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | X-ray structure of succinylglutamate desuccinalase from Vibrio Parahaemolyticus (RIMD 2210633) at the resolution 2.3 A, Northeast Structural Genomics Target Vpr14
To be Published
|
|
1AZW
 
 | | PROLINE IMINOPEPTIDASE FROM XANTHOMONAS CAMPESTRIS PV. CITRI | | Descriptor: | PROLINE IMINOPEPTIDASE | | Authors: | Medrano, F.J, Alonso, J, Garcia, J.L, Romero, A, Bode, W, Gomis-Ruth, F.X. | | Deposit date: | 1997-11-22 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of proline iminopeptidase from Xanthomonas campestris pv. citri: a prototype for the prolyl oligopeptidase family.
EMBO J., 17, 1998
|
|
