4EGG
 
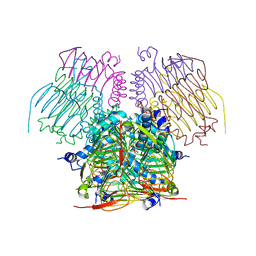 | | Computationally Designed Self-assembling tetrahedron protein, T310 | | 分子名称: | GLYCEROL, Putative acetyltransferase SACOL2570 | | 著者 | Sawaya, M.R, King, N.P, Sheffler, W, Baker, D, Yeates, T.O. | | 登録日 | 2012-03-30 | | 公開日 | 2012-05-30 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Computational design of self-assembling protein nanomaterials with atomic level accuracy.
Science, 336, 2012
|
|
1QUS
 
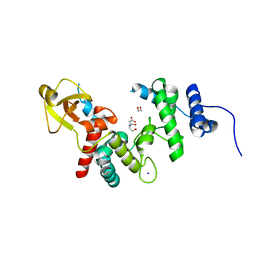 | | 1.7 A RESOLUTION STRUCTURE OF THE SOLUBLE LYTIC TRANSGLYCOSYLASE SLT35 FROM ESCHERICHIA COLI | | 分子名称: | 1,2-ETHANEDIOL, BICINE, LYTIC MUREIN TRANSGLYCOSYLASE B, ... | | 著者 | van Asselt, E.J, Dijkstra, A.J, Kalk, K.H, Takacs, B, Keck, W, Dijkstra, B.W. | | 登録日 | 1999-07-03 | | 公開日 | 1999-09-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of Escherichia coli lytic transglycosylase Slt35 reveals a lysozyme-like catalytic domain with an EF-hand.
Structure Fold.Des., 7, 1999
|
|
1R02
 
 | |
4EP1
 
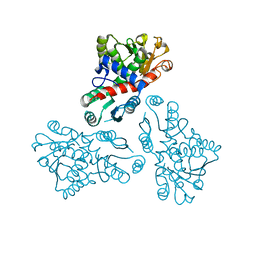 | | Crystal structure of anabolic ornithine carbamoyltransferase from Bacillus anthracis | | 分子名称: | Ornithine carbamoyltransferase | | 著者 | Shabalin, I.G, Mikolajczak, K, Stam, J, Winsor, J, Shuvalova, L, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2012-04-16 | | 公開日 | 2012-04-25 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.25 Å) | | 主引用文献 | Crystal structures of anabolic ornithine carbamoyltransferase from Bacillus anthracis
To be Published
|
|
3NIL
 
 | | The structure of UBR box (RDAA) | | 分子名称: | ACETATE ION, E3 ubiquitin-protein ligase UBR1, Peptide RDAA, ... | | 著者 | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | 登録日 | 2010-06-16 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3TMA
 
 | | Crystal structure of TrmN from Thermus thermophilus | | 分子名称: | PHOSPHATE ION, methyltransferase | | 著者 | Fislage, M, Roovers, M, Tuszynska, I, Bujnicki, J.M, Droogmans, L, Versees, W. | | 登録日 | 2011-08-31 | | 公開日 | 2012-03-14 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Crystal structures of the tRNA:m2G6 methyltransferase Trm14/TrmN from two domains of life.
Nucleic Acids Res., 40, 2012
|
|
3NIK
 
 | | The structure of UBR box (REAA) | | 分子名称: | E3 ubiquitin-protein ligase UBR1, Peptide REAA, ZINC ION | | 著者 | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | 登録日 | 2010-06-16 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIS
 
 | | The structure of UBR box (native2) | | 分子名称: | ACETATE ION, E3 ubiquitin-protein ligase UBR1, ZINC ION | | 著者 | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | 登録日 | 2010-06-16 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
1QEW
 
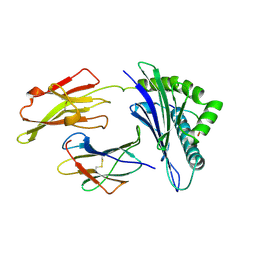 | | HUMAN CLASS I HISTOCOMPATIBILITY ANTIGEN (HLA-A 0201) COMPLEX WITH A NONAMERIC PEPTIDE FROM MELANOMA-ASSOCIATED ANTIGEN 3 (RESIDUES 271-279) | | 分子名称: | PROTEIN (BETA-2-MICROGLOBULIN), PROTEIN (HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, B-35 B* 3501 ALPHA CHAIN), ... | | 著者 | Orth, P, Alings, C, Saenger, W, Ziegler, A. | | 登録日 | 1999-04-02 | | 公開日 | 2003-11-18 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | HUMAN CLASS I HISTOCOMPATIBILITY ANTIGEN (HLA-A 0201)
COMPLEX WITH A NONAMERIC PEPTIDE FROM MELANOMA-ASSOCIATED
ANTIGEN 3 (RESIDUES 271-279)
To be Published
|
|
4ED3
 
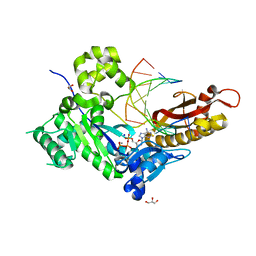 | | Human DNA polymerase eta - DNA ternary complex: AT crystal at pH 7.5 (Na+ HEPES) with 1 Ca2+ ion | | 分子名称: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), ... | | 著者 | Nakamura, T, Zhao, Y, Yang, W. | | 登録日 | 2012-03-26 | | 公開日 | 2012-07-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.791 Å) | | 主引用文献 | Watching DNA polymerase eta make a phosphodiester bond
Nature, 487, 2012
|
|
3TUG
 
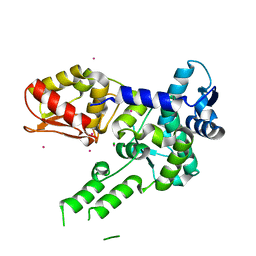 | | Crystal structure of the HECT domain of ITCH E3 ubiquitin ligase | | 分子名称: | CHLORIDE ION, E3 ubiquitin-protein ligase Itchy homolog, UNKNOWN ATOM OR ION | | 著者 | Dong, A, Dobrovetsky, E, Xue, S, Butler, C, Wernimont, A, Walker, J.R, Tempel, W, Dhe-Paganon, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Tong, Y, Structural Genomics Consortium (SGC) | | 登録日 | 2011-09-16 | | 公開日 | 2011-10-12 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | Crystal structure of the HECT domain of ITCH E3 ubiquitin ligase
To be Published
|
|
3TUH
 
 | |
1QZ1
 
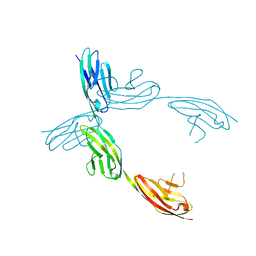 | | Crystal Structure of the Ig 1-2-3 fragment of NCAM | | 分子名称: | Neural cell adhesion molecule 1, 140 kDa isoform | | 著者 | Soroka, V, Kolkova, K, Kastrup, J.S, Diederichs, K, Breed, J, Kiselyov, V.V, Poulsen, F.M, Larsen, I.K, Welte, W, Berezin, V, Bock, E, Kasper, C. | | 登録日 | 2003-09-15 | | 公開日 | 2003-11-04 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure and interactions of NCAM Ig1-2-3 suggest a novel zipper mechanism for homophilic adhesion
Structure, 11, 2003
|
|
1QZC
 
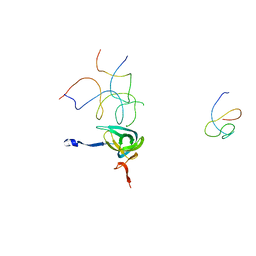 | | Coordinates of S12, SH44, LH69 and SRL separately fitted into the cryo-EM map of EF-Tu ternary complex (GDP.Kirromycin) bound 70S ribosome | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S12 | | 著者 | Valle, M, Zavialov, A, Li, W, Stagg, S.M, Sengupta, J, Nielsen, R.C, Nissen, P, Harvey, S.C, Ehrenberg, M, Frank, J. | | 登録日 | 2003-09-16 | | 公開日 | 2003-11-04 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (9 Å) | | 主引用文献 | Incorporation of Aminoacyl-tRNA into the Ribosome as seen by Cryo-electron Microscopy
Nat.Struct.Biol., 10, 2003
|
|
1QZB
 
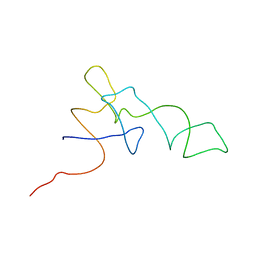 | | Coordinates of the A-site tRNA model fitted into the cryo-EM map of 70S ribosome in the pre-translocational state | | 分子名称: | Phe-tRNA | | 著者 | Valle, M, Zavialov, A, Li, W, Stagg, S.M, Sengupta, J, Nielsen, R.C, Nissen, P, Harvey, S.C, Ehrenberg, M, Frank, J. | | 登録日 | 2003-09-16 | | 公開日 | 2003-11-04 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (9 Å) | | 主引用文献 | Incorporation of Aminoacyl-tRNA into the Ribosome as seen by Cryo-electron Microscopy
Nat.Struct.Biol., 10, 2003
|
|
8HVJ
 
 | |
4EC6
 
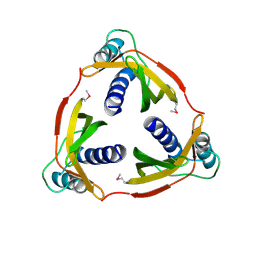 | | Ntf2-like, potential transfer protein TraM from Gram-positive conjugative plasmid pIP501 | | 分子名称: | Putative uncharacterized protein | | 著者 | Goessweiner-Mohr, N, Grumet, L, Pavkov-Keller, T, Wang, M, Keller, W. | | 登録日 | 2012-03-26 | | 公開日 | 2012-12-05 | | 最終更新日 | 2013-02-06 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The 2.5 A Structure of the Enterococcus Conjugation Protein TraM resembles VirB8 Type IV Secretion Proteins.
J.Biol.Chem., 288, 2013
|
|
1QVR
 
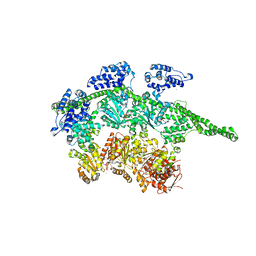 | | Crystal Structure Analysis of ClpB | | 分子名称: | ClpB protein, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PLATINUM (II) ION | | 著者 | Lee, S, Sowa, M.E, Watanabe, Y, Sigler, P.B, Chiu, W, Yoshida, M, Tsai, F.T.F. | | 登録日 | 2003-08-28 | | 公開日 | 2003-10-21 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The Structure of ClpB: A Molecular Chaperone that Rescues Proteins from an Aggregated State
Cell(Cambridge,Mass.), 115, 2003
|
|
8TA5
 
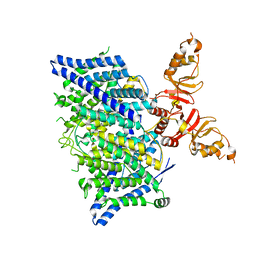 | | Title: Cryo-EM structure of the human CLC-2 chloride channel transmembrane domain with asymmetric C-terminal | | 分子名称: | Chloride channel protein 2 | | 著者 | Xu, M, Neelands, T, Powers, A.S, Liu, Y, Miller, S, Pintilie, G, Du Bois, J, Dror, R.O, Chiu, W, Maduke, M. | | 登録日 | 2023-06-26 | | 公開日 | 2024-01-31 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (2.76 Å) | | 主引用文献 | CryoEM structures of the human CLC-2 voltage-gated chloride channel reveal a ball-and-chain gating mechanism.
Elife, 12, 2024
|
|
4EF4
 
 | | Crystal structure of STING CTD complex with c-di-GMP | | 分子名称: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CALCIUM ION, Transmembrane protein 173 | | 著者 | Ouyang, S, Ru, H, Shaw, N, Jiang, Y, Niu, F, Zhu, Y, Qiu, W, Li, Y, Liu, Z.-J. | | 登録日 | 2012-03-29 | | 公開日 | 2012-05-16 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.147 Å) | | 主引用文献 | Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding
Immunity, 36, 2012
|
|
3U4A
 
 | | From soil to structure: a novel dimeric family 3-beta-glucosidase isolated from compost using metagenomic analysis | | 分子名称: | CALCIUM ION, JMB19063, beta-D-glucopyranose, ... | | 著者 | McAndrew, R.P, Park, J.I, Reindl, W, Friedland, G.D, D'haeseleer, P, Northen, T, Sale, K.L, Simmons, B.A, Adams, P.D. | | 登録日 | 2011-10-07 | | 公開日 | 2013-04-10 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.196 Å) | | 主引用文献 | From soil to structure: a novel dimeric family 3-beta--glucosidase isolated from compost using metagenomic analysis
To be Published
|
|
8HZ5
 
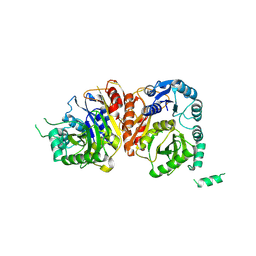 | |
3TZK
 
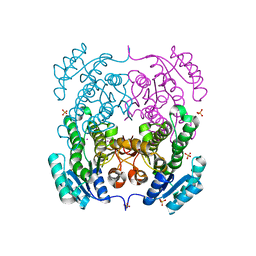 | | Crystal structure of 3-ketoacyl-(acyl-carrier-protein) reductase (FabG)(G92A) from Vibrio cholerae | | 分子名称: | 3-oxoacyl-[acyl-carrier protein] reductase, SULFATE ION, UNKNOWN ATOM OR ION | | 著者 | Hou, J, Chruszcz, M, Zheng, H, Grabowski, M, Domagalski, M, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-09-27 | | 公開日 | 2011-10-19 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Dissecting the Structural Elements for the Activation of beta-Ketoacyl-(Acyl Carrier Protein) Reductase from Vibrio cholerae.
J.Bacteriol., 198, 2015
|
|
8TA4
 
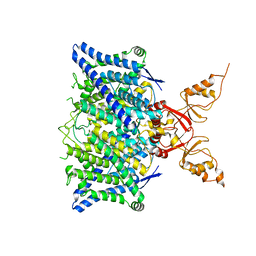 | | Cryo-EM structure of the human CLC-2 chloride channel transmembrane domain with symmetric C-terminal | | 分子名称: | CHLORIDE ION, Chloride channel protein 2 | | 著者 | Xu, M, Neelands, T, Powers, A.S, Liu, Y, Miller, S, Pintilie, G, Du Bois, J, Dror, R.O, Chiu, W, Maduke, M. | | 登録日 | 2023-06-26 | | 公開日 | 2024-01-31 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (2.75 Å) | | 主引用文献 | CryoEM structures of the human CLC-2 voltage-gated chloride channel reveal a ball-and-chain gating mechanism.
Elife, 12, 2024
|
|
1QZD
 
 | | EF-Tu.kirromycin coordinates fitted into the cryo-EM map of EF-Tu ternary complex (GDP.Kirromycin) bound 70S ribosome | | 分子名称: | Elongation factor Tu | | 著者 | Valle, M, Zavialov, A, Li, W, Stagg, S.M, Sengupta, J, Nielsen, R.C, Nissen, P, Harvey, S.C, Ehrenberg, M, Frank, J. | | 登録日 | 2003-09-16 | | 公開日 | 2003-11-04 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (10 Å) | | 主引用文献 | Incorporation of Aminoacyl-tRNA into the Ribosome as seen by Cryo-electron Microscopy
Nat.Struct.Biol., 10, 2003
|
|
