7GGM
 
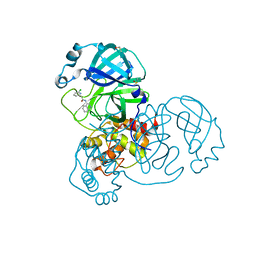 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-afd4d4fd-2 (Mpro-x12677) | | 分子名称: | 2-(6-chloropyridin-2-yl)-N-(isoquinolin-4-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.839 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
6HOY
 
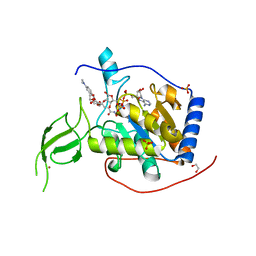 | |
3WOK
 
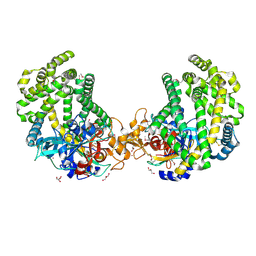 | | Crystal structure of the DAP BII (Space) | | 分子名称: | GLYCEROL, ZINC ION, dipeptidyl aminopeptidase BII | | 著者 | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | 登録日 | 2013-12-29 | | 公開日 | 2014-09-03 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | S46 peptidases are the first exopeptidases to be members of clan PA
SCI REP, 4, 2014
|
|
7GGO
 
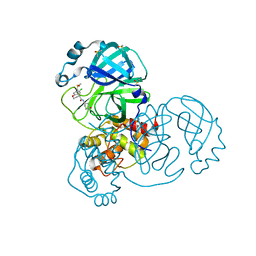 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ALP-POS-8b8a49e1-4 (Mpro-x12682) | | 分子名称: | (4R)-6-chloro-N-[(4R)-2-oxopiperidin-4-yl]-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.687 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GGP
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with VLA-UCB-29506327-1 (Mpro-x12686) | | 分子名称: | (1'M,4S)-6-chloro-1'-(isoquinolin-4-yl)-2,3-dihydrospiro[[1]benzopyran-4,4'-imidazolidine]-2',5'-dione, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.902 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GH2
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-090737b9-1 (Mpro-x12735) | | 分子名称: | (4R)-6-chloro-N-(isoquinolin-4-yl)-N-propanoyl-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GH4
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDJ-MED-00c1612e-1 (Mpro-x12777) | | 分子名称: | 2-(3-chlorophenyl)-N-(6-methoxyisoquinolin-4-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GH5
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with TRY-UNI-714-12 (Mpro-x2908) | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(3-chlorophenyl)-N'-(4-methylpyridin-3-yl)urea | | 著者 | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | 登録日 | 2023-08-11 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
4MUY
 
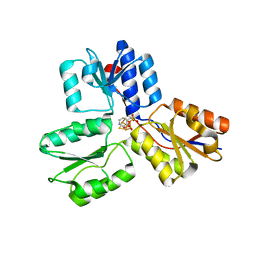 | | IspH in complex with pyridin-4-ylmethyl diphosphate | | 分子名称: | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, FE3-S4 CLUSTER, pyridin-4-ylmethyl trihydrogen diphosphate | | 著者 | Span, I, Wang, K, Song, Y, Eisenreich, W, Bacher, A, Oldfield, E, Groll, M. | | 登録日 | 2013-09-23 | | 公開日 | 2014-06-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Insights into the Binding of Pyridines to the Iron-Sulfur Enzyme IspH.
J.Am.Chem.Soc., 136, 2014
|
|
6HOJ
 
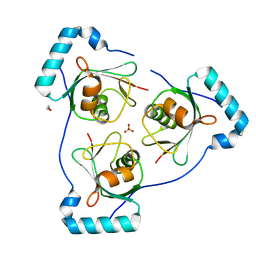 | | Structure of Beclin1 LIR motif bound to GABARAP | | 分子名称: | 1,2-ETHANEDIOL, Beclin-1,Gamma-aminobutyric acid receptor-associated protein, SULFATE ION | | 著者 | Mouilleron, S, Birgisdottir, A.B, Bhujbal, Z, Wirth, M, Sjottem, E, Evjen, G, Zhang, W, Lee, R, O'Reilly, N, Tooze, S, Lamark, T, Johansen, T. | | 登録日 | 2018-09-17 | | 公開日 | 2019-02-27 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs.
Autophagy, 15, 2019
|
|
3WVJ
 
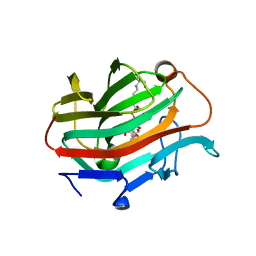 | | The crystal structure of native glycosidic hydrolase | | 分子名称: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucanase | | 著者 | Chen, C.C, Huang, J.W, Zhao, P, Ko, T.P, Huang, C.H, Chan, H.C, Huang, Z, Liu, W, Cheng, Y.S, Liu, J.R, Guo, R.T. | | 登録日 | 2014-05-22 | | 公開日 | 2015-06-24 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural analyses and yeast production of the beta-1,3-1,4-glucanase catalytic module encoded by the licB gene of Clostridium thermocellum.
Enzyme.Microb.Technol., 71, 2015
|
|
4V06
 
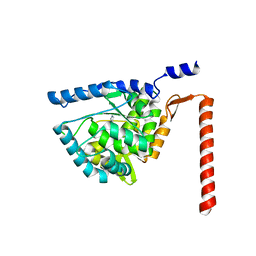 | | Crystal structure of human tryptophan hydroxylase 2 (TPH2), catalytic domain | | 分子名称: | FE (III) ION, IMIDAZOLE, TRYPTOPHAN 5-HYDROXYLASE 2 | | 著者 | Kopec, J, Oberholzer, A, Fitzpatrick, F, Newman, J, Tallant, C, Kiyani, W, Shrestha, L, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Yue, W.W. | | 登録日 | 2014-09-11 | | 公開日 | 2014-10-15 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.63 Å) | | 主引用文献 | Crystal Structure of Human Tryptophane Hydroxylase 2 (Tph2), Catalytic Domain
To be Published
|
|
4MMH
 
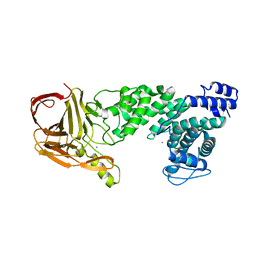 | | Crystal structure of heparan sulfate lyase HepC from Pedobacter heparinus | | 分子名称: | CALCIUM ION, Heparinase III protein | | 著者 | Maruyama, Y, Nakamichi, Y, Mikami, B, Murata, K, Hashimoto, W. | | 登録日 | 2013-09-09 | | 公開日 | 2014-01-29 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal Structure of Pedobacter heparinus Heparin Lyase Hep III with the Active Site in a Deep Cleft
Biochemistry, 53, 2014
|
|
6HRF
 
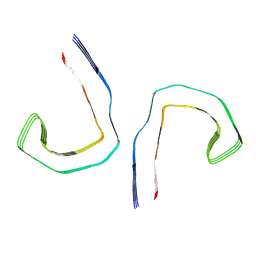 | | Straight filament from sporadic Alzheimer's disease brain | | 分子名称: | Microtubule-associated protein tau | | 著者 | Falcon, B, Zhang, W, Schweighauser, M, Murzin, A.G, Vidal, R, Garringer, H.J, Ghetti, B, Scheres, S.H.W, Goedert, M. | | 登録日 | 2018-09-26 | | 公開日 | 2018-10-10 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Tau filaments from multiple cases of sporadic and inherited Alzheimer's disease adopt a common fold.
Acta Neuropathol., 136, 2018
|
|
3WEA
 
 | |
3WKF
 
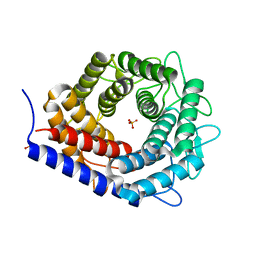 | | Crystal structure of cellobiose 2-epimerase | | 分子名称: | CHLORIDE ION, Cellobiose 2-epimerase, PHOSPHATE ION | | 著者 | Fujiwara, T, Saburi, W, Tanaka, I, Yao, M. | | 登録日 | 2013-10-21 | | 公開日 | 2013-12-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.743 Å) | | 主引用文献 | Structural Insights into the Epimerization of beta-1,4-Linked Oligosaccharides Catalyzed by Cellobiose 2-Epimerase, the Sole Enzyme Epimerizing Non-anomeric Hydroxyl Groups of Unmodified Sugars
J.Biol.Chem., 289, 2014
|
|
4MOU
 
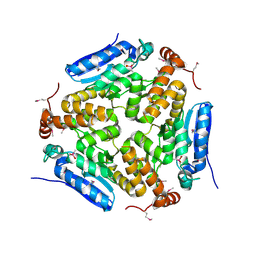 | | Crystal Structure of an Enoyl-CoA Hydratase/Isomerase Family Member, NYSGRC target 028282 | | 分子名称: | CACODYLATE ION, Enoyl-CoA hydratase/isomerase family protein | | 著者 | Chapman, H.C, Cooper, D.R, Geffken, K.T, Cymborowski, M.T, Osinski, T, Stead, M, Hillerich, B, Ahmed, M, Bonanno, J.B, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2013-09-12 | | 公開日 | 2013-09-25 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Crystal Structure of an Enoyl-CoA Hydratase/Isomerase Family Member, NYSGRC target 028282
To be Published
|
|
4MMI
 
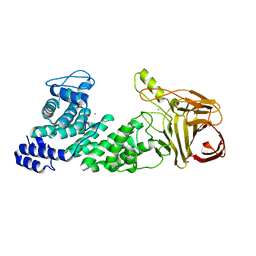 | | Crystal structure of heparan sulfate lyase HepC mutant from Pedobacter heparinus | | 分子名称: | CALCIUM ION, Heparinase III protein | | 著者 | Maruyama, Y, Nakamichi, Y, Mikami, B, Murata, K, Hashimoto, W. | | 登録日 | 2013-09-09 | | 公開日 | 2014-01-29 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal Structure of Pedobacter heparinus Heparin Lyase Hep III with the Active Site in a Deep Cleft
Biochemistry, 53, 2014
|
|
4MPL
 
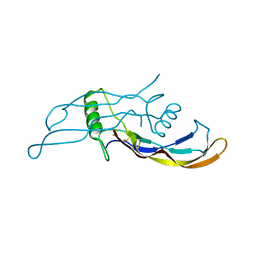 | |
6HYO
 
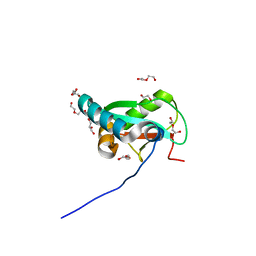 | | Structure of ULK1 LIR motif bound to GABARAP | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Mouilleron, S, Wirth, M, Zhang, W, O'Reilly, N, Tooze, S, Johansen, T, Razi, M, Nyoni, L, Joshi, D. | | 登録日 | 2018-10-22 | | 公開日 | 2019-05-08 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.07 Å) | | 主引用文献 | Molecular determinants regulating selective binding of autophagy adapters and receptors to ATG8 proteins.
Nat Commun, 10, 2019
|
|
4NQ0
 
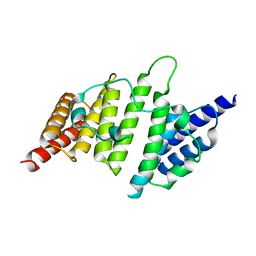 | | Structural insights into yeast histone chaperone Hif1: a scaffold protein recruiting protein complexes to core histones | | 分子名称: | HAT1-interacting factor 1 | | 著者 | Liu, H, Zhang, M, He, W, Zhu, Z, Teng, M, Gao, Y, Niu, L. | | 登録日 | 2013-11-23 | | 公開日 | 2014-07-16 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural insights into yeast histone chaperone Hif1: a scaffold protein recruiting protein complexes to core histones
Biochem.J., 462, 2014
|
|
4NPL
 
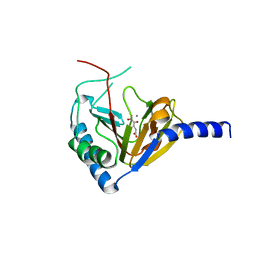 | |
3WOJ
 
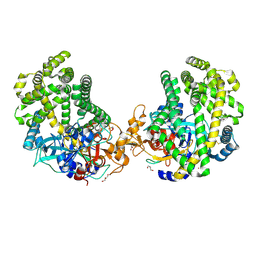 | | Crystal structure of the DAP BII | | 分子名称: | GLYCEROL, ZINC ION, dipeptidyl aminopeptidase BII | | 著者 | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | 登録日 | 2013-12-29 | | 公開日 | 2014-09-03 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | S46 peptidases are the first exopeptidases to be members of clan PA
SCI REP, 4, 2014
|
|
3WTP
 
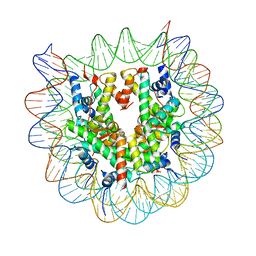 | | Crystal Structure of the heterotypic nucleosome containing human CENP-A and H3.3 | | 分子名称: | DNA (146-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | 著者 | Arimura, Y, Shirayama, K, Horikoshi, N, Fujita, R, Kagawa, W, Fukagawa, T, Almouzni, G, Kurumizaka, H. | | 登録日 | 2014-04-14 | | 公開日 | 2014-12-03 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | Crystal structure and stable property of the cancer-associated heterotypic nucleosome containing CENP-A and H3.3
Sci Rep, 4, 2014
|
|
3WU4
 
 | | Oxidized-form structure of E.coli Lon Proteolytic domain | | 分子名称: | Lon protease, SULFATE ION | | 著者 | Nishii, W, Kukimoto-Niino, M, Terada, T, Shirouzu, M, Muramatsu, T, Yokoyama, S. | | 登録日 | 2014-04-22 | | 公開日 | 2014-11-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | A redox switch shapes the Lon protease exit pore to facultatively regulate proteolysis.
Nat. Chem. Biol., 11, 2015
|
|
