1G26
 
 | |
1OBB
 
 | | alpha-glucosidase A, AglA, from Thermotoga maritima in complex with maltose and NAD+ | | Descriptor: | ALPHA-GLUCOSIDASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Lodge, J.A, Maier, T, Liebl, W, Hoffmann, V, Strater, N. | | Deposit date: | 2003-01-29 | | Release date: | 2003-05-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Thermotoga Maritima Alpha-Glucosidase Agla Defines a New Clan of Nad+-Dependent Glycosidases
J.Biol.Chem., 278, 2003
|
|
1G2A
 
 | | THE CRYSTAL STRUCTURE OF E.COLI PEPTIDE DEFORMYLASE COMPLEXED WITH ACTINONIN | | Descriptor: | ACTINONIN, NICKEL (II) ION, POLYPEPTIDE DEFORMYLASE | | Authors: | Clements, J.M, Beckett, P, Brown, A, Catlin, C, Lobell, M, Palan, S, Thomas, W, Whittaker, M, Baker, P.J, Rodgers, H.F, Barynin, V, Rice, D.W, Hunter, M.G. | | Deposit date: | 2000-10-18 | | Release date: | 2001-10-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Antibiotic activity and characterization of BB-3497, a novel peptide deformylase inhibitor.
Antimicrob.Agents Chemother., 45, 2001
|
|
6BKX
 
 | | Novel Modes of Inhibition of Wild-Type IDH1: Direct Covalent Modification of His315 with Cmpd1 | | Descriptor: | (6aS,7S,9R,10aS)-7,10a-dimethyl-8-oxo-2-(phenylamino)-5,6,6a,7,8,9,10,10a-octahydrobenzo[h]quinazoline-9-carbonitrile, CALCIUM ION, ISOCITRIC ACID, ... | | Authors: | Jakob, C.G, Qiu, W. | | Deposit date: | 2017-11-09 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Novel Modes of Inhibition of Wild-Type Isocitrate Dehydrogenase 1 (IDH1): Direct Covalent Modification of His315.
J. Med. Chem., 61, 2018
|
|
6BSH
 
 | | Structure of HIV-1 RT complexed with RNA/DNA hybrid in the RNA hydrolysis mode | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, CALCIUM ION, DNA (5'-D(*GP*TP*AP*TP*GP*CP*CP*AP*CP*TP*AP*GP*TP*TP*AP*TP*TP*GP*TP*GP*GP*CP*C)-3'), ... | | Authors: | Tian, L, Kim, M, Yang, W. | | Deposit date: | 2017-12-03 | | Release date: | 2018-01-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.649 Å) | | Cite: | Structure of HIV-1 reverse transcriptase cleaving RNA in an RNA/DNA hybrid.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1NMN
 
 | | Structure of yqgF from Escherichia coli, a hypothetical protein | | Descriptor: | Hypothetical protein yqgF | | Authors: | Galkin, A, Sarikaya, E, Krajewski, W, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2003-01-10 | | Release date: | 2004-03-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of yqgF from Escherichia coli, a hypothetical protein
To be Published
|
|
3K3S
 
 | | Crystal structure of altronate hydrolase (fragment 1-84) from Shigella Flexneri. | | Descriptor: | ACETATE ION, Altronate hydrolase, CHLORIDE ION, ... | | Authors: | Hou, J, Chruszcz, M, Xu, X, Le, B, Zimmerman, M.D, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-10-04 | | Release date: | 2009-10-27 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of altronate hydrolase (fragment 1-84) from Shigella Flexneri.
To be Published
|
|
1G3J
 
 | | CRYSTAL STRUCTURE OF THE XTCF3-CBD/BETA-CATENIN ARMADILLO REPEAT COMPLEX | | Descriptor: | BETA-CATENIN ARMADILLO REPEAT REGION, TCF3-CBD (CATENIN BINDING DOMAIN) | | Authors: | Graham, T.A, Weaver, C, Mao, F, Kimelman, D, Xu, W. | | Deposit date: | 2000-10-24 | | Release date: | 2000-12-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a beta-catenin/Tcf complex.
Cell(Cambridge,Mass.), 103, 2000
|
|
5HU2
 
 | |
3K57
 
 | | Crystal structure of E.coli Pol II-normal DNA-dATP ternary complex | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (5'-D(*G*TP*AP*TP*GP*TP*AP*CP*GP*CP*TP*AP*GP*GP*CP*AP*CP*G)-3'), DNA (5'-D(*GP*TP*GP*CP*CP*TP*AP*GP*CP*GP*TP*AP*(DOC))-3'), ... | | Authors: | Yang, W, Wang, F. | | Deposit date: | 2009-10-06 | | Release date: | 2010-02-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural insight into translesion synthesis by DNA Pol II
Cell(Cambridge,Mass.), 139, 2009
|
|
1NPR
 
 | | CRYSTAL STRUCTURE OF AQUIFEX AEOLICUS NUSG IN C222(1) | | Descriptor: | Transcription antitermination protein nusG | | Authors: | Knowlton, J.R, Bubunenko, M, Andrykovitch, M, Guo, W, Routzhan, K.M, Waugh, D.S, Court, D.L, Ji, X. | | Deposit date: | 2003-01-18 | | Release date: | 2003-03-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | A Spring-Loaded State of NusG in Its Functional Cycle Is Suggested by X-ray Crystallography and Supported by
Site-Directed Mutants
Biochemistry, 42, 2003
|
|
3QGL
 
 | |
1NQV
 
 | | Crystal Structure of Lumazine Synthase from Aquifex aeolicus in Complex with Inhibitor: 5-nitroso-6-ribityl-amino-2,4(1H,3H)pyrimidinedione | | Descriptor: | 5-NITROSO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION | | Authors: | Zhang, X, Meining, W, Cushman, M, Haase, I, Fischer, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2003-01-23 | | Release date: | 2004-01-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A structure-based model of the reaction catalyzed by lumazine synthase from Aquifex aeolicus.
J.Mol.Biol., 328, 2003
|
|
1NTH
 
 | | Crystal structure of the methanosarcina barkeri monomethylamine methyltransferase (MTMB) | | Descriptor: | Monomethylamine methyltransferase mtmB1 | | Authors: | Hao, B, Gong, W, Ferguson, T.K, James, C.M, Krzycki, J.A, Chan, M.K. | | Deposit date: | 2003-01-30 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A new UAG-encoded residue in the structure of a methanogen methyltransferase
Science, 296, 2002
|
|
5HZ0
 
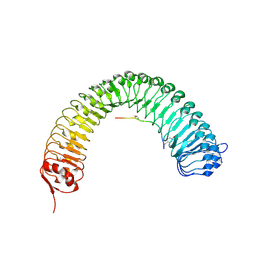 | | Plant peptide hormone receptor RGFR1 in complex with RGF2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ASP-PTR-TRP-LYS-PRO-ARG-HIS-HIS-PRO-HYP-ARG-ASN-ASN, ... | | Authors: | Song, W, Han, Z, Chai, J. | | Deposit date: | 2016-02-02 | | Release date: | 2017-04-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Plant Receptor
To Be Published
|
|
5HI0
 
 | | The Substrate Binding Mode and Chemical Basis of a Reaction Specificity Switch in Oxalate Decarboxylase | | Descriptor: | COBALT (II) ION, OXALATE ION, Oxalate decarboxylase OxdC, ... | | Authors: | Zhu, W, Easthon, L.M, Reinhardt, L.A, Tu, C, Cohen, S.E, Silverman, D.N, Allen, K.N, Richards, N.G.J. | | Deposit date: | 2016-01-11 | | Release date: | 2016-04-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Substrate Binding Mode and Molecular Basis of a Specificity Switch in Oxalate Decarboxylase.
Biochemistry, 55, 2016
|
|
5HZQ
 
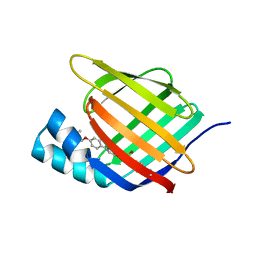 | | Crystal structure of cellular retinoic acid binding protein 2 (CRABP2)-aryl fluorosulfate covalent conjugate | | Descriptor: | 4'-[(3,6,9,12-tetraoxapentadec-14-yn-1-yl)oxy][1,1'-biphenyl]-4-yl sulfurofluoridate, Cellular retinoic acid-binding protein 2, GLYCEROL | | Authors: | Chen, W, Mortenson, D.E, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2016-02-02 | | Release date: | 2016-06-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Arylfluorosulfates Inactivate Intracellular Lipid Binding Protein(s) through Chemoselective SuFEx Reaction with a Binding Site Tyr Residue.
J.Am.Chem.Soc., 138, 2016
|
|
3QKG
 
 | |
3K8V
 
 | |
6BSR
 
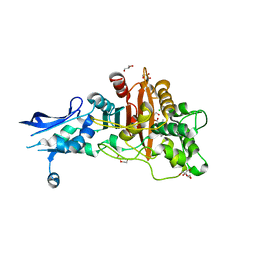 | | Crystal structure of penicillin-binding protein 4 (PBP4) from Enterococcus faecalis in the benzylpenicillin bound form. | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Moon, T.M, D'Andrea, E.D, Peti, W, Page, R. | | Deposit date: | 2017-12-04 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
5HIQ
 
 | |
3K9A
 
 | | Crystal Structure of HIV gp41 with MPER | | Descriptor: | HIV glycoprotein gp41 | | Authors: | Shi, W, Han, D, Habte, H, Cho, M, Chance, M.R. | | Deposit date: | 2009-10-15 | | Release date: | 2010-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of HIV gp41 with the membrane-proximal external region
J.Biol.Chem., 285, 2010
|
|
6BUI
 
 | |
5HLT
 
 | | Crystal structure of pyrene- and phenanthrene-modified DNA in complex with the BpuJ1 endonuclease binding domain | | Descriptor: | DNA (5'-D(*GP*YPY*TP*AP*CP*CP*CP*GP*TP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*AP*CP*GP*GP*GP*TP*AP*YPY*C)-3'), Restriction endonuclease R.BpuJI | | Authors: | Probst, M, Aeschimann, W, Chau, T.-T.-H, Langenegger, S.M, Stocker, A, Haener, R. | | Deposit date: | 2016-01-15 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.672 Å) | | Cite: | Structural insight into DNA-assembled oligochromophores: crystallographic analysis of pyrene- and phenanthrene-modified DNA in complex with BpuJI endonuclease.
Nucleic Acids Res., 44, 2016
|
|
3J3X
 
 | |
