3RHY
 
 | | Crystal structure of the dimethylarginine dimethylaminohydrolase adduct with 4-chloro-2-hydroxymethylpyridine | | Descriptor: | (4-chloropyridin-2-yl)methanol, N(G),N(G)-dimethylarginine dimethylaminohydrolase | | Authors: | Monzingo, A.F, Johnson, C.M, Ke, Z, Yoon, D.-W, Linsky, T.W, Guo, H, Fast, W, Robertus, J.D. | | Deposit date: | 2011-04-12 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | On the mechanism of dimethylarginine dimethylaminohydrolase inactivation by 4-halopyridines.
J.Am.Chem.Soc., 133, 2011
|
|
2BU2
 
 | | crystal structures of human pyruvate dehydrogenase kinase 2 containing physiological and synthetic ligands | | Descriptor: | 4-({(2R,5S)-2,5-DIMETHYL-4-[(2R)-3,3,3-TRIFLUORO-2-HYDROXY-2-METHYLPROPANOYL]PIPERAZIN-1-YL}CARBONYL)BENZONITRILE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Knoechel, T.R, Tucker, A.D, Robinson, C.M, Phillips, C, Taylor, W, Bungay, P.J, Kasten, S.A, Roche, T.E, Brown, D.G. | | Deposit date: | 2005-06-08 | | Release date: | 2006-02-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Regulatory Roles of the N-Terminal Domain Based on Crystal Structures of Human Pyruvate Dehydrogenase Kinase 2 Containing Physiological and Synthetic Ligands.
Biochemistry, 45, 2006
|
|
7WPA
 
 | | SARS-CoV-2 Omicron Variant SPIKE trimer complexed with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S, Xu, E. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
7WP9
 
 | | SARS-CoV-2 Omicron Variant SPIKE trimer, all RBDs down | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S, Xu, E. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
7WPB
 
 | | SARS-CoV-2 Omicron Variant RBD complexed with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, CHLORIDE ION, ... | | Authors: | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S, Xu, E. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
7WPC
 
 | | The second RBD of SARS-CoV-2 Omicron Variant in complexed with RBD-ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S, Xu, E. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
7WPD
 
 | | SARS-CoV-2 Omicron Variant S Trimer complexed with one JMB2002 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Anti-Fab nanobody, ... | | Authors: | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S.J, Xu, H.E. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
4IH4
 
 | | Crystal structure of Arabidopsis DWARF14 orthologue, AtD14 | | Descriptor: | AT3g03990/T11I18_10 | | Authors: | Zhou, X.E, Zhao, L.-H, Wu, Z.-S, Yi, W, Li, S, Li, Y, Xu, Y, Xu, T.-H, Liu, Y, Chen, R.-Z, Kovach, A, Kang, Y, Hou, L, He, Y, Zhang, C, Melcher, K, Xu, H.E. | | Deposit date: | 2012-12-18 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of two phytohormone signal-transducing alpha / beta hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14.
Cell Res., 23, 2013
|
|
2W36
 
 | | Structures of endonuclease V with DNA reveal initiation of deaminated adenine repair | | Descriptor: | 5'-D(*CP*GP*AP*TP*CP*TP*GP*TP*AP*GP*CP)-3', 5'-D(*GP*CP*BRUP*AP*CP*IP*GP*AP*BRUP*CP*GP)-3', ENDONUCLEASE V | | Authors: | Dalhus, B, Arvai, A.S, Rosnes, I, Olsen, O.E, Backe, P.H, Alseth, I, Gao, H, Cao, W, Tainer, J.A, Bjoras, M. | | Deposit date: | 2008-11-06 | | Release date: | 2009-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Endonuclease V with DNA Reveal Initiation of Deaminated Adenine Repair.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3R0H
 
 | | Structure of INAD PDZ45 in complex with NG2 peptide | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Inactivation-no-after-potential D protein, ... | | Authors: | Wei, Z, Liu, W, Zhang, M. | | Deposit date: | 2011-03-08 | | Release date: | 2011-11-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The INAD scaffold is a dynamic, redox-regulated modulator of signaling in the Drosophila eye
Cell(Cambridge,Mass.), 145, 2011
|
|
2VSI
 
 | | Synthesis of CDP-activated ribitol for teichoic acid precursors in Streptococcus pneumoniae | | Descriptor: | 2-C-METHYL-D-ERYTHRITOL 4-PHOSPHATE CYTIDYLYLTRANSFERASE, 4-AMINO-1-{5-O-[(R)-HYDROXY(PHOSPHONOOXY)PHOSPHORYL]-ALPHA-D-ARABINOFURANOSYL}PYRIMIDIN-2(1H)-ONE, CALCIUM ION | | Authors: | Baur, S, Marles-Wright, J, Buckenmaier, S, Lewis, R.J, Vollmer, W. | | Deposit date: | 2008-04-23 | | Release date: | 2008-12-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Synthesis of Cdp-Activated Ribitol for Teichoic Acid Precursors in Streptococcus Pneumoniae.
J.Bacteriol., 191, 2009
|
|
2BDH
 
 | | Human Kallikrein 4 complex with zinc and p-aminobenzamidine | | Descriptor: | Kallikrein-4, P-AMINO BENZAMIDINE, ZINC ION | | Authors: | Debela, M, Bode, W, Goettig, P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-10-20 | | Release date: | 2006-10-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of human tissue kallikrein 4: activity modulation by a specific zinc binding site.
J.Mol.Biol., 362, 2006
|
|
2BJG
 
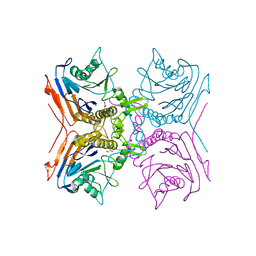 | | Crystal Structure of Conjugated Bile Acid Hydrolase from Clostridium perfringens in Complex with Reaction Products Taurine and Deoxycholate | | Descriptor: | 1,2-ETHANEDIOL, CHOLOYLGLYCINE HYDROLASE | | Authors: | Rossocha, M, Schultz-Heienbrok, R, Von Moeller, H, Coleman, J.P, Saenger, W. | | Deposit date: | 2005-02-02 | | Release date: | 2005-05-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conjugated Bile Acid Hydrolase is a Tetrameric N-Terminal Thiol Hydrolase with Specific Recognition of its Cholyl But not of its Tauryl Product
Biochemistry, 44, 2005
|
|
2X3T
 
 | | Glutaraldehyde-crosslinked wheat germ agglutinin isolectin 1 crystal soaked with a synthetic glycopeptide | | Descriptor: | 2-acetamido-1-O-carbamoyl-2-deoxy-alpha-D-glucopyranose, AGGLUTININ ISOLECTIN 1, D-ALPHA-AMINOBUTYRIC ACID, ... | | Authors: | Schwefel, D, Maierhofer, C, Wittmann, V, Diederichs, K, Welte, W. | | Deposit date: | 2010-01-26 | | Release date: | 2010-06-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.749 Å) | | Cite: | Structural Basis of Multivalent Binding to Wheat Germ Agglutinin.
J.Am.Chem.Soc., 132, 2010
|
|
2X6O
 
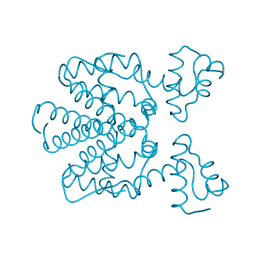 | | Tet Repressor class D in complex with 7-chlor-2-cyano-iso- tetracycline | | Descriptor: | (4S,4AS,6S,8AS)-6-[(1S)-7-CHLORO-4-HYDROXY-1-METHYL-3-OXO-1,3-DIHYDRO-2-BENZOFURAN-1-YL]-4-(DIMETHYLAMINO)-3,8A-DIHYDROXY-1,8-DIOXO-1,4,4A,5,6,7,8,8A-OCTAHYDRONAPHTHALENE-2-CARBONITRILE, CHLORIDE ION, TETRACYCLINE REPRESSOR PROTEIN CLASS D | | Authors: | Volkers, G, Hinrichs, W. | | Deposit date: | 2010-02-18 | | Release date: | 2011-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Recognition of Drug Degradation Products by Target Proteins: Isotetracycline Binding to Tet Repressor.
J.Med.Chem., 54, 2011
|
|
2BQ2
 
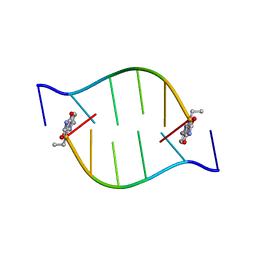 | | Solution Structure of the DNA Duplex ACGCGU-NA with a 2' Amido-Linked Nalidixic Acid Residue at the 3' Terminal Nucleotide | | Descriptor: | 5'-D(*AP*CP*GP*CP*GP*2AU)-3', NALIDIXIC ACID | | Authors: | Siegmund, K, Maheshwary, S, Narayanan, S, Connors, W, Richert, M. | | Deposit date: | 2005-04-26 | | Release date: | 2006-08-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular details of quinolone-DNA interactions: solution structure of an unusually stable DNA duplex with covalently linked nalidixic acid residues and non-covalent complexes derived from it.
Nucleic Acids Res., 33, 2005
|
|
1B85
 
 | | LIGNIN PEROXIDASE | | Descriptor: | CALCIUM ION, Ligninase H8, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Blodig, W, Doyle, W.A, Smith, A.T, Piontek, K. | | Deposit date: | 1999-02-05 | | Release date: | 1999-02-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of pristine and oxidatively processed lignin peroxidase expressed in Escherichia coli and of the W171F variant that eliminates the redox active tryptophan 171. Implications for the reaction mechanism.
J.Mol.Biol., 305, 2001
|
|
8P6I
 
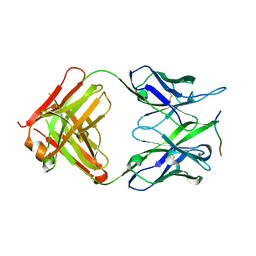 | | Crystal structure of the 139H2 Fab fragment bound to Muc1 peptide epitope | | Descriptor: | 139H2 HC, 139H2 LC, Mucin-1 | | Authors: | Beugelink, J.W, Peng, W, Siborova, M, Pronker, M.F, Snijder, J, Janssen, B.J.C. | | Deposit date: | 2023-05-26 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Reverse-engineering the anti-MUC1 antibody 139H2 by mass spectrometry-based de novo sequencing.
Life Sci Alliance, 7, 2024
|
|
2BU6
 
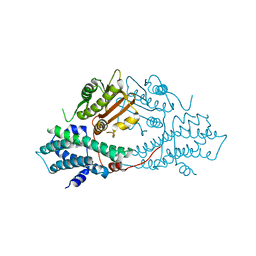 | | crystal structures of human pyruvate dehydrogenase kinase 2 containing physiological and synthetic ligands | | Descriptor: | (N-{4-[(ETHYLANILINO)SULFONYL]-2-METHYLPHENYL}-3,3,3-TRIFLUORO-2-HYDROXY-2-METHYLPROPANAMIDE, PYRUVATE DEHYDROGENASE KINASE ISOENZYME 2 | | Authors: | Knoechel, T.R, Tucker, A.D, Robinson, C.M, Phillips, C, Taylor, W, Bungay, P.J, Kasten, S.A, Roche, T.E, Brown, D.G. | | Deposit date: | 2005-06-08 | | Release date: | 2006-02-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Regulatory Roles of the N-Terminal Domain Based on Crystal Structures of Human Pyruvate Dehydrogenase Kinase 2 Containing Physiological and Synthetic Ligands.
Biochemistry, 45, 2006
|
|
2WW7
 
 | |
8P2I
 
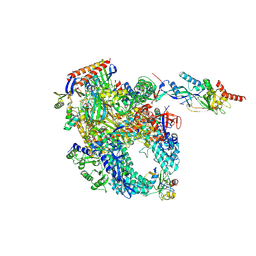 | | Cryo-EM structure of Pyrococcus furiosus apo form RNA polymerase contracted clamp conformation with Spt4/5 | | Descriptor: | DNA-directed RNA polymerase subunit Rpo10, DNA-directed RNA polymerase subunit Rpo11, DNA-directed RNA polymerase subunit Rpo12, ... | | Authors: | Tarau, D.M, Reichelt, R, Heiss, F.B, Pilsl, M, Hausner, W, Engel, C, Grohmann, D. | | Deposit date: | 2023-05-16 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of archaeal RNA polymerase transcription elongation and Spt4/5 recruitment.
Nucleic Acids Res., 52, 2024
|
|
2WHV
 
 | | CRYSTAL STRUCTURE OF MOUSE CADHERIN-23 EC1-2 (ALL CATION BINDING SITES OCCUPIED BY CALCIUM) | | Descriptor: | CADHERIN-23, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Sotomayor, M, Weihofen, W, Gaudet, R, Corey, D.P. | | Deposit date: | 2009-05-07 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural Determinants of Cadherin-23 Function in Hearing and Deafness.
Neuron, 66, 2010
|
|
8PIC
 
 | | Cryo EM structure of the type 3B polymorph of alpha-synuclein at low pH. | | Descriptor: | Alpha-synuclein, CHLORIDE ION | | Authors: | Frey, L, Qureshi, B.M, Kwiatkowski, W, Rhyner, D, Greenwald, J, Riek, R. | | Deposit date: | 2023-06-21 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | On the pH-dependence of alpha-synuclein amyloid polymorphism and the role of secondary nucleation in seed-based amyloid propagation
Elife, 2023
|
|
2C1C
 
 | | Structural basis of the resistance of an insect carboxypeptidase to plant protease inhibitors | | Descriptor: | CARBOXYPEPTIDASE B, YTTRIUM ION, ZINC ION | | Authors: | Bayes, A, Comellas-Bigler, M, Rodriguez de la Vega, M, Maskos, K, Bode, W, Aviles, F.X, Jongsma, M.A, Beekwilder, J, Vendrell, J. | | Deposit date: | 2005-09-12 | | Release date: | 2005-10-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of the Resistance of an Insect Carboxypeptidase to Plant Protease Inhibitors.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2WJH
 
 | | Structure and function of the FeoB G-domain from Methanococcus jannaschii | | Descriptor: | CITRATE ANION, FERROUS IRON TRANSPORT PROTEIN B HOMOLOG, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Koester, S, Wehner, M, Herrmann, C, Kuehlbrandt, W, Yildiz, O. | | Deposit date: | 2009-05-26 | | Release date: | 2009-07-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Function of the Feob G-Domain from Methanococcus Jannaschii
J.Mol.Biol., 392, 2009
|
|
