4U6S
 
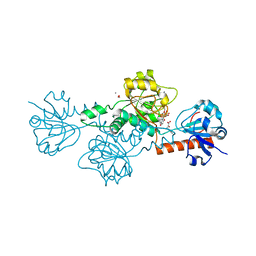 | | CtBP1 in complex with substrate phenylpyruvate | | Descriptor: | 3-PHENYLPYRUVIC ACID, C-terminal-binding protein 1, CALCIUM ION, ... | | Authors: | Hilbert, B.J, Morris, B.L, Ellis, K.C, Paulsen, J.L, Schiffer, C.A, Grossman, S.R, Royer Jr, W.E. | | Deposit date: | 2014-07-29 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Design of a High Affinity Inhibitor to Human CtBP.
Acs Chem.Biol., 10, 2015
|
|
1YHU
 
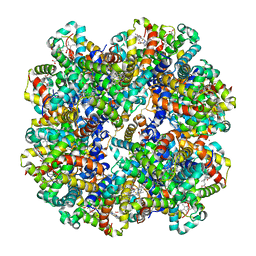 | | Crystal structure of Riftia pachyptila C1 hemoglobin reveals novel assembly of 24 subunits. | | Descriptor: | Giant hemoglobins B chain, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Flores, J.F, Fisher, C.R, Carney, S.L, Green, B.N, Freytag, J.K, Schaeffer, S.W, Royer, W.E. | | Deposit date: | 2005-01-10 | | Release date: | 2005-02-08 | | Last modified: | 2013-03-06 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Sulfide binding is mediated by zinc ions discovered in the crystal structure of a hydrothermal vent tubeworm hemoglobin.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
3SDH
 
 | |
3SNP
 
 | |
4BG2
 
 | | X-ray Crystal Structure of PatF from Prochloron didemni | | Descriptor: | PATF | | Authors: | Bent, A.F, Koehnke, J, Houssen, W.E, Smith, M.C.M, Jaspars, M, Naismith, J.H. | | Deposit date: | 2013-03-22 | | Release date: | 2013-04-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structure of Patf from Prochloron Didemni.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4BS9
 
 | | Structure of the heterocyclase TruD | | Descriptor: | TRUD, ZINC ION | | Authors: | Koehnke, J, Bent, A.F, Zollman, D, Smith, K, Houssen, W.E, Zhu, X, Mann, G, Lebl, T, Scharff, R, Shirran, S, Botting, C.H, Jaspars, M, Schwarz-Linek, U, Naismith, J.H. | | Deposit date: | 2013-06-09 | | Release date: | 2013-11-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Cyanobactin Heterocyclase Enzyme: A Processive Adenylase that Operates with a Defined Order of Reaction.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
4AKT
 
 | | PatG macrocyclase in complex with peptide | | Descriptor: | SUBSTRATE ANALOGUE, THIAZOLINE OXIDASE/SUBTILISIN-LIKE PROTEASE | | Authors: | Koehnke, J, Bent, A, Houssen, W.E, Zollman, D, Morawitz, F, Shirran, S, Vendome, J, Nneoyiegbe, A.F, Trembleau, L, Botting, C.H, Smith, M.C.M, Jaspars, M, Naismith, J.H. | | Deposit date: | 2012-02-28 | | Release date: | 2012-07-18 | | Last modified: | 2013-11-06 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | The Mechanism of Patellamide Macrocyclization Revealed by the Characterization of the Patg Macrocyclase Domain.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4AKS
 
 | | PatG macrocyclase domain | | Descriptor: | THIAZOLINE OXIDASE/SUBTILISIN-LIKE PROTEASE | | Authors: | Koehnke, J, Bent, A, Houssen, W.E, Zollman, D, Morawitz, F, Shirran, S, Vendome, J, Nneoyiegbe, A.F, Trembleau, L, Botting, C.H, Smith, M.C.M, Jaspars, M, Naismith, J.H. | | Deposit date: | 2012-02-28 | | Release date: | 2012-07-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The Mechanism of Patellamide Macrocyclization Revealed by the Characterization of the Patg Macrocyclase Domain.
Nat.Struct.Mol.Biol., 19, 2012
|
|
5JRM
 
 | | Crystal Structure of a Xylanase at 1.56 Angstroem resolution | | Descriptor: | Endo-1,4-beta-xylanase, GLYCEROL, SULFATE ION | | Authors: | Gomez, S, Payne, A.M, Savko, M, Fox, G.C, Shepard, W.E, Fernandez, F.J, Vega, M.C. | | Deposit date: | 2016-05-06 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural and functional characterization of a highly stable endo-beta-1,4-xylanase from Fusarium oxysporum and its development as an efficient immobilized biocatalyst.
Biotechnol Biofuels, 9, 2016
|
|
5JRN
 
 | | Crystal Structure of a Xylanase in Complex with a Monosaccharide at 2.84 Angstroem resolution | | Descriptor: | Endo-1,4-beta-xylanase, GLYCEROL, methyl beta-D-xylopyranoside | | Authors: | Gomez, S, Payne, A.M, Savko, M, Fox, G.C, Shepard, W.E, Fernandez, F.J, Vega, M.C. | | Deposit date: | 2016-05-06 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.841 Å) | | Cite: | Structural and functional characterization of a highly stable endo-beta-1,4-xylanase from Fusarium oxysporum and its development as an efficient immobilized biocatalyst.
Biotechnol Biofuels, 9, 2016
|
|
5LAC
 
 | |
5LAK
 
 | | Ligand-bound structure of Cavally Virus 3CL Protease | | Descriptor: | 3Cl Protease, BEZ-TYR-TYR-ASN-ECC Peptide inhibitor, PENTAETHYLENE GLYCOL, ... | | Authors: | Kanitz, M, Heine, A, Diederich, W.E. | | Deposit date: | 2016-06-14 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Structural basis for catalysis and substrate specificity of a 3C-like cysteine protease from a mosquito mesonivirus.
Virology, 533, 2019
|
|
5KEG
 
 | | Crystal structure of APOBEC3A in complex with a single-stranded DNA | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA (5'-D(*TP*TP*CP*TP*T)-3'), ... | | Authors: | Kouno, T, Hilbert, B.J, Silvas, T, Royer, W.E, Matsuo, H, Schiffer, C.A. | | Deposit date: | 2016-06-09 | | Release date: | 2017-05-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of APOBEC3A bound to single-stranded DNA reveals structural basis for cytidine deamination and specificity.
Nat Commun, 8, 2017
|
|
2Z8A
 
 | | Ligand Migration and Binding in The Dimeric Hemoglobin of Scapharca Inaequivalvis: I25W with CO Bound to HEME and in the Presence of 3 Atoms of XE | | Descriptor: | CARBON MONOXIDE, Globin-1, PHOSPHATE ION, ... | | Authors: | Knapp, J.E, Royer Jr, W.E, Nienhaus, K, Palladino, P, Nienhaus, G.U. | | Deposit date: | 2007-09-04 | | Release date: | 2007-11-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Ligand Migration and Binding in the Dimeric Hemoglobin of Scapharca inaequivalvis
Biochemistry, 46, 2007
|
|
6CDF
 
 | | Human CtBP1 (28-378) | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, C-terminal-binding protein 1, CALCIUM ION, ... | | Authors: | Royer, W.E, Bellesis, A.G. | | Deposit date: | 2018-02-08 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Assembly of human C-terminal binding protein (CtBP) into tetramers.
J. Biol. Chem., 293, 2018
|
|
6CDR
 
 | | Human CtBP1 (28-378) | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, C-terminal-binding protein 1, ... | | Authors: | Royer, W.E, Bellesis, A.G. | | Deposit date: | 2018-02-09 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Assembly of human C-terminal binding protein (CtBP) into tetramers.
J. Biol. Chem., 293, 2018
|
|
2N9C
 
 | | NRAS Isoform 5 | | Descriptor: | GTPase NRas | | Authors: | Markowitz, J, Mal, T.K, Yuan, C, Courtney, N.B, Patel, M, Stiff, A.R, Blachly, J, Walker, C, Eisfeld, A, de la Chapelle, A, Carson III, W.E. | | Deposit date: | 2015-11-13 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of NRAS isoform 5.
Protein Sci., 25, 2016
|
|
2OIP
 
 | |
6BQG
 
 | | Crystal structure of 5-HT2C in complex with ergotamine | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5-hydroxytryptamine receptor 2C,Soluble cytochrome b562, Ergotamine | | Authors: | Peng, Y, McCorvy, J.D, Harpsoe, K, Lansu, K, Yuan, S, Popov, P, Qu, L, Pu, M, Che, T, Nikolajse, L.F, Huang, X.P, Wu, Y, Shen, L, Bjorn-Yoshimoto, W.E, Ding, K, Wacker, D, Han, G.W, Cheng, J, Katritch, V, Jensen, A.A, Hanson, M.A, Zhao, S, Gloriam, D.E, Roth, B.L, Stevens, R.C, Liu, Z. | | Deposit date: | 2017-11-27 | | Release date: | 2018-02-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 5-HT2C Receptor Structures Reveal the Structural Basis of GPCR Polypharmacology.
Cell, 172, 2018
|
|
2Z85
 
 | | Ligand Migration and Binding in The Dimeric Hemoglobin of Scapharca Inaequivalvis: M37F Unliganded | | Descriptor: | Globin-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Knapp, J.E, Royer Jr, W.E, Nienhaus, K, Palladino, P, Nienhaus, G.U. | | Deposit date: | 2007-09-02 | | Release date: | 2007-11-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Ligand Migration and Binding in the Dimeric Hemoglobin of Scapharca inaequivalvis
Biochemistry, 46, 2007
|
|
6BQH
 
 | | Crystal structure of 5-HT2C in complex with ritanserin | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5-hydroxytryptamine receptor 2C,Soluble cytochrome b562, 6-(2-{4-[bis(4-fluorophenyl)methylidene]piperidin-1-yl}ethyl)-7-methyl-5H-[1,3]thiazolo[3,2-a]pyrimidin-5-one, ... | | Authors: | Peng, Y, McCorvy, J.D, Harpsoe, K, Lansu, K, Yuan, S, Popov, P, Qu, L, Pu, M, Che, T, Nikolajse, L.F, Huang, X.P, Wu, Y, Shen, L, Bjorn-Yoshimoto, W.E, Ding, K, Wacker, D, Han, G.W, Cheng, J, Katritch, V, Jensen, A.A, Hanson, M.A, Zhao, S, Gloriam, D.E, Roth, B.L, Stevens, R.C, Liu, Z. | | Deposit date: | 2017-11-27 | | Release date: | 2018-02-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 5-HT2C Receptor Structures Reveal the Structural Basis of GPCR Polypharmacology.
Cell, 172, 2018
|
|
2LHB
 
 | |
6E0I
 
 | | Crystal structure of Glucokinase in complex with compound 72 | | Descriptor: | 1-{4-[5-({3-[(2-methylpyridin-3-yl)oxy]-5-[(pyridin-2-yl)sulfanyl]pyridin-2-yl}amino)-1,2,4-thiadiazol-3-yl]piperidin-1 -yl}ethan-1-one, DIMETHYL SULFOXIDE, Glucokinase, ... | | Authors: | Hinklin, R.J, Baer, B.R, Boyd, S.A, Chicarelli, M.D, Condroski, K.R, DeWolf, W.E, Fischer, J, Frank, M, Hingorani, G.P, Lee, P.A, Neitzel, N.A, Pratt, S.A, Singh, A, Sullivan, F.X, Turner, T, Voegtli, W.C, Wallace, E.M, Williams, L, Aicher, T.D. | | Deposit date: | 2018-07-06 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and preclinical development of AR453588 as an anti-diabetic glucokinase activator.
Bioorg.Med.Chem., 28, 2020
|
|
6E0E
 
 | | Crystal structure of Glucokinase in complex with compound 6 | | Descriptor: | 2-({2-[(4-methyl-1,3-thiazol-2-yl)amino]pyridin-3-yl}oxy)benzonitrile, Glucokinase, alpha-D-glucopyranose | | Authors: | Hinklin, R.J, Baer, B.R, Boyd, S.A, Chicarelli, M.D, Condroski, K.R, DeWolf, W.E, Fischer, J, Frank, M, Hingorani, G.P, Lee, P.A, Neitzel, N.A, Pratt, S.A, Singh, A, Sullivan, F.X, Turner, T, Voegtli, W.C, Wallace, E.M, Williams, L, Aicher, T.D. | | Deposit date: | 2018-07-06 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery and preclinical development of AR453588 as an anti-diabetic glucokinase activator.
Bioorg.Med.Chem., 28, 2020
|
|
2MCX
 
 | | Solid-state NMR structure of piscidin 3 in aligned 1:1 phosphatidylethanolamine/phosphoglycerol lipid bilayers | | Descriptor: | Piscidin-3 | | Authors: | Fu, R, Tian, Y, Perrin Jr, B.S, Grant, C.V, Wieczorek, W.E, Pastor, R.W, Cotten, M.L. | | Deposit date: | 2013-08-27 | | Release date: | 2014-01-22 | | Last modified: | 2024-10-30 | | Method: | SOLID-STATE NMR | | Cite: | High-resolution structures and orientations of antimicrobial peptides piscidin 1 and piscidin 3 in fluid bilayers reveal tilting, kinking, and bilayer immersion.
J.Am.Chem.Soc., 136, 2014
|
|
