1YOE
 
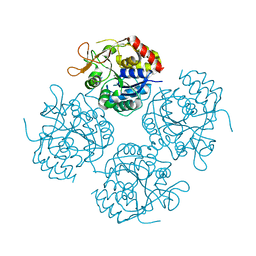 | |
5K1S
 
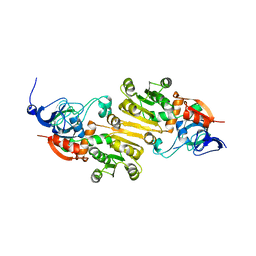 | | crystal structure of AibC | | Descriptor: | Oxidoreductase, zinc-binding dehydrogenase family, ZINC ION | | Authors: | Bock, T, Mueller, R, Blankenfeldt, W. | | Deposit date: | 2016-05-18 | | Release date: | 2016-08-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of AibC, a reductase involved in alternative de novo isovaleryl coenzyme A biosynthesis in Myxococcus xanthus.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
8H1D
 
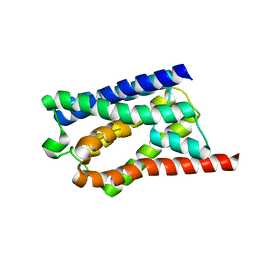 | | Solid-state NMR Structure of Aquaporin Z in its Native Cellular Membranes | | Descriptor: | Aquaporin Z | | Authors: | Xie, H, Zhao, Y, Zhao, W, Chen, Y, Liu, M, Yang, J. | | Deposit date: | 2022-10-02 | | Release date: | 2022-11-09 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Solid-state NMR structure determination of a membrane protein in E. coli cellular inner membrane.
Sci Adv, 9, 2023
|
|
4H2M
 
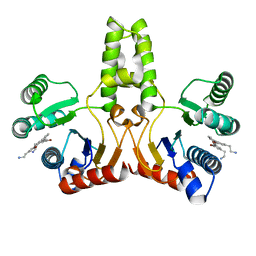 | | Structure of E. coli undecaprenyl diphosphate synthase in complex with BPH-1408 | | Descriptor: | 2,2'-{benzene-1,3-diylbis[ethyne-2,1-diyl(5-bromobenzene-3,1-diyl)]}diethanamine, Undecaprenyl pyrophosphate synthase | | Authors: | Zhu, W, Oldfield, E. | | Deposit date: | 2012-09-12 | | Release date: | 2012-12-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Antibacterial drug leads targeting isoprenoid biosynthesis.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5LNF
 
 | | Solution NMR structure of farnesylated PEX19, C-terminal domain | | Descriptor: | FARNESYL, Peroxisomal biogenesis factor 19 | | Authors: | Emmanouilidis, L, Schuetz, U, Tripsianes, K, Madl, T, Radke, J, Rucktaeschel, R, Wilmanns, M, Schliebs, W, Erdmann, R, Sattler, M. | | Deposit date: | 2016-08-04 | | Release date: | 2017-03-15 | | Last modified: | 2019-09-11 | | Method: | SOLUTION NMR | | Cite: | Allosteric modulation of peroxisomal membrane protein recognition by farnesylation of the peroxisomal import receptor PEX19.
Nat Commun, 8, 2017
|
|
5M04
 
 | | Structure of ObgE from Escherichia coli | | Descriptor: | GTPase ObgE/CgtA, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Gkekas, S, Singh, R.K, Versees, W. | | Deposit date: | 2016-10-03 | | Release date: | 2017-03-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and biochemical analysis of Escherichia coli ObgE, a central regulator of bacterial persistence.
J. Biol. Chem., 292, 2017
|
|
4H3A
 
 | |
5LPE
 
 | | Kallikrein-related peptidase 10 complex with Zn2+ | | Descriptor: | Kallikrein-10, SULFATE ION, ZINC ION | | Authors: | Goettig, P, Debela, M, Magdolen, V, Bode, W, Brandstetter, H. | | Deposit date: | 2016-08-12 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis for the Zn2+ inhibition of the zymogen-like kallikrein-related peptidase 10.
Biol.Chem., 397, 2016
|
|
5LPF
 
 | | Kallikrein-related peptidase 10 | | Descriptor: | Kallikrein-10, SULFATE ION | | Authors: | Goettig, P, Debela, M, Magdolen, V, Bode, W, Brandstetter, H. | | Deposit date: | 2016-08-12 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the Zn2+ inhibition of the zymogen-like kallikrein-related peptidase 10.
Biol.Chem., 397, 2016
|
|
5M3K
 
 | | A multi-component acyltransferase PhlABC from Pseudomonas protegens | | Descriptor: | 2,4-diacetylphloroglucinol biosynthesis protein PhlB, 2,4-diacetylphloroglucinol biosynthesis protein PhlC, PhlA, ... | | Authors: | Pavkov-Keller, T, Schmidt, N.G, Kroutil, W, Gruber, K. | | Deposit date: | 2016-10-15 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Structure and Catalytic Mechanism of a Bacterial Friedel-Crafts Acylase.
Chembiochem, 20, 2019
|
|
1UXW
 
 | | CRYSTAL STRUCTURE OF HLA-B*2709 COMPLEXED WITH THE LATENT MEMBRANE PROTEIN 2 PEPTIDE (LMP2) OF EPSTEIN-BARR VIRUS | | Descriptor: | BETA-2-MICROGLOBULIN, GENE TERMINAL PROTEIN (MEMBRANE PROTEIN LMP-2A/LMP-2B), GLYCEROL, ... | | Authors: | Hulsmeyer, M, Kozerski, C, Fiorillo, M.T, Sorrentino, R, Saenger, W, Ziegler, A, Uchanska-Ziegler, B. | | Deposit date: | 2004-03-01 | | Release date: | 2004-11-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Allele-Dependent Similarity between Viral and Self-Peptide Presentation by Hla-B27 Subtypes
J.Biol.Chem., 280, 2005
|
|
5J1V
 
 | | Crystal structure of human CLK1 in complex with pyrido[3,4-g]quinazoline derivative ZW29 (compound 13) | | Descriptor: | Dual specificity protein kinase CLK1, GLYCEROL, pyrido[3,4-g]quinazoline-2,10-diamine | | Authors: | Chaikuad, A, Esvan, Y.J, Zeinyeh, W, Boibessot, T, Nauton, L, Thery, V, Loaec, N, Meijer, L, Giraud, F, Moreau, P, Anizon, F, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-03-29 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Discovery of pyrido[3,4-g]quinazoline derivatives as CMGC family protein kinase inhibitors: Design, synthesis, inhibitory potency and X-ray co-crystal structure.
Eur.J.Med.Chem., 118, 2016
|
|
5J2Y
 
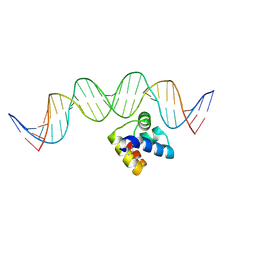 | | Molecular insight into the regulatory mechanism of the quorum-sensing repressor RsaL in Pseudomonas aeruginosa | | Descriptor: | DNA (26-MER), Regulatory protein | | Authors: | Zhao, J, Gan, J, Zhang, J, Kang, H, Kong, W, Zhu, M, Li, F, Song, Y, Qin, J, Liang, H. | | Deposit date: | 2016-03-30 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Pseudomonas aeruginosa RsaL bound to promoter DNA reaffirms its role as a global regulator involved in quorum-sensing.
Nucleic Acids Res., 45, 2017
|
|
5JDV
 
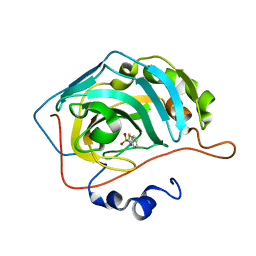 | | Human carbonic anhydrase II (F131W) complexed with benzo[d]thiazole-2-sulfonamide | | Descriptor: | 1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Fox, J.M, Kang, K, Sastry, M, Sherman, W, Sankaran, B, Zwart, P.H, Whitesides, G.M. | | Deposit date: | 2016-04-17 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Water-Restructuring Mutations Can Reverse the Thermodynamic Signature of Ligand Binding to Human Carbonic Anhydrase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
8JHG
 
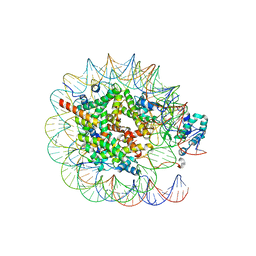 | | Native SUV420H1 bound to 167-bp nucleosome | | Descriptor: | DNA (160-MER), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | Authors: | Lin, F, Li, W. | | Deposit date: | 2023-05-23 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural basis of nucleosomal H4K20 recognition and methylation by SUV420H1 methyltransferase.
Cell Discov, 9, 2023
|
|
5JGF
 
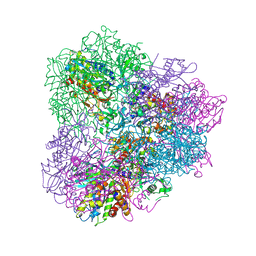 | | Crystal structure of mApe1 | | Descriptor: | Vacuolar aminopeptidase 1, ZINC ION | | Authors: | Noda, N.N, Adachi, W, Inagaki, F. | | Deposit date: | 2016-04-20 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Basis for Receptor-Mediated Selective Autophagy of Aminopeptidase I Aggregates
Cell Rep, 16, 2016
|
|
5JCB
 
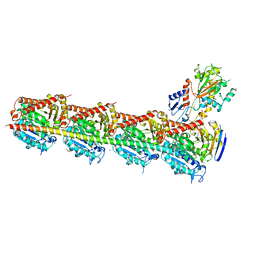 | | Microtubule depolymerizing agent podophyllotoxin derivative YJTSF1 | | Descriptor: | (5R,5aR,8aS,9R)-9-[(4H-1,2,4-triazol-3-yl)sulfanyl]-5-(3,4,5-trimethoxyphenyl)-5,8,8a,9-tetrahydro-2H-furo[3',4':6,7]naphtho[2,3-d][1,3]dioxol-6(5aH)-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Guan, Z, Zhao, W, Yin, P. | | Deposit date: | 2016-04-14 | | Release date: | 2017-09-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insights into the Inhibition of Tubulin by the Antitumor Agent 4 beta-(1,2,4-triazol-3-ylthio)-4-deoxypodophyllotoxin.
ACS Chem. Biol., 12, 2017
|
|
1ML1
 
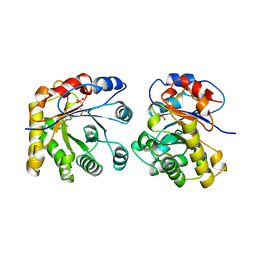 | | PROTEIN ENGINEERING WITH MONOMERIC TRIOSEPHOSPHATE ISOMERASE: THE MODELLING AND STRUCTURE VERIFICATION OF A SEVEN RESIDUE LOOP | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Thanki, N, Zeelen, J.P, Mathieu, M, Jaenicke, R, Abagyan, R.A, Wierenga, R, Schliebs, W. | | Deposit date: | 1996-09-27 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Protein engineering with monomeric triosephosphate isomerase (monoTIM): the modelling and structure verification of a seven-residue loop.
Protein Eng., 10, 1997
|
|
5JEH
 
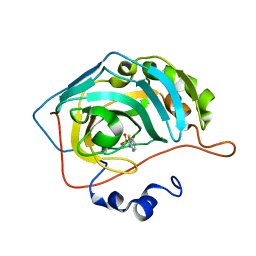 | | Human carbonic anhydrase II (L198A) complexed with benzo[d]thiazole-2-sulfonamide | | Descriptor: | 1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Fox, J.M, Kang, K, Sastry, M, Sherman, W, Sankaran, B, Zwart, P.H, Whitesides, G.M. | | Deposit date: | 2016-04-18 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Water-Restructuring Mutations Can Reverse the Thermodynamic Signature of Ligand Binding to Human Carbonic Anhydrase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5JG6
 
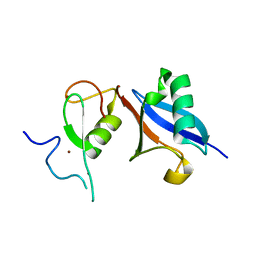 | | APC11-Ubv shows role of noncovalent RING-Ubiquitin interactions in processive multiubiquitination and Ubiquitin chain elongation by APC/C | | Descriptor: | Anaphase-promoting complex subunit 11, Polyubiquitin-B, ZINC ION | | Authors: | Brown, N.G, Zhang, W, Yu, S, Miller, D.J, Sidhu, S.S, Schulman, B.A. | | Deposit date: | 2016-04-19 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.0013 Å) | | Cite: | Dual RING E3 Architectures Regulate Multiubiquitination and Ubiquitin Chain Elongation by APC/C.
Cell, 165, 2016
|
|
5M1M
 
 | |
4H4D
 
 | | IspH in complex with (E)-4-amino-3-methylbut-2-enyl diphosphate | | Descriptor: | (2E)-4-amino-3-methylbut-2-en-1-yl trihydrogen diphosphate, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IRON/SULFUR CLUSTER | | Authors: | Span, I, Eisenreich, W, Jauch, J, Bacher, A, Groll, M. | | Deposit date: | 2012-09-17 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structures of Fluoro, Amino, and Thiol Inhibitors Bound to the [Fe(4) S(4) ] Protein IspH.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
4H8E
 
 | |
1OVO
 
 | | CRYSTALLOGRAPHIC REFINEMENT OF JAPANESE QUAIL OVOMUCOID, A KAZAL-TYPE INHIBITOR, AND MODEL BUILDING STUDIES OF COMPLEXES WITH SERINE PROTEASES | | Descriptor: | OVOMUCOID THIRD DOMAIN | | Authors: | Weber, E, Papamokos, E, Bode, W, Huber, R, Kato, I, Laskowskijunior, M. | | Deposit date: | 1982-01-18 | | Release date: | 1982-05-26 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic refinement of Japanese quail ovomucoid, a Kazal-type inhibitor, and model building studies of complexes with serine proteases.
J.Mol.Biol., 158, 1982
|
|
1MQS
 
 | |
