5HCN
 
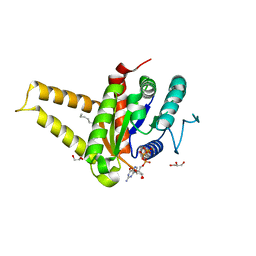 | | GPN-loop GTPase Npa3 in complex with GMPPCP | | Descriptor: | GLYCEROL, GPN-loop GTPase 1, LAURIC ACID, ... | | Authors: | Niesser, J, Wagner, F.R, Kostrewa, D, Muehlbacher, W, Cramer, P. | | Deposit date: | 2016-01-04 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of GPN-Loop GTPase Npa3 and Implications for RNA Polymerase II Assembly.
Mol.Cell.Biol., 36, 2015
|
|
5ZYR
 
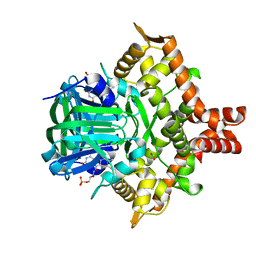 | | Crystal structure of the reductase (C1) component of p-hydroxyphenylacetate 3-hydroxylase (HPAH) from Acinetobacter baumannii | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, p-hydroxyphenylacetate 3-hydroxylase, ... | | Authors: | Oonanant, W, Phongsak, T, Sucharitakul, J, Chaiyen, P, Yuvaniyama, J. | | Deposit date: | 2018-05-28 | | Release date: | 2019-06-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.20001316 Å) | | Cite: | Crystal structure of the reductase (C1) component of p-hydroxyphenylacetate 3-hydroxylase (HPAH) from Acinetobacter baumannii
To Be Published
|
|
5HCI
 
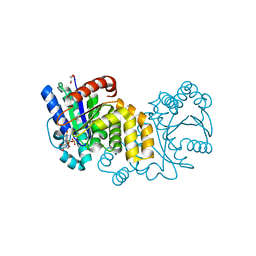 | | GPN-loop GTPase Npa3 in complex with GDP | | Descriptor: | GLYCEROL, GPN-loop GTPase 1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Niesser, J, Wagner, F.R, Kostrewa, D, Muehlbacher, W, Cramer, P. | | Deposit date: | 2016-01-04 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of GPN-Loop GTPase Npa3 and Implications for RNA Polymerase II Assembly.
Mol.Cell.Biol., 36, 2015
|
|
1ECA
 
 | |
1ES7
 
 | |
3QT2
 
 | | Structure of a cytokine ligand-receptor complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Interleukin-5, Interleukin-5 receptor subunit alpha, ... | | Authors: | Mueller, T.D, Patino, E, Kotzsch, A, Saremba, S, Nickel, J, Schmitz, W, Sebald, W. | | Deposit date: | 2011-02-22 | | Release date: | 2012-02-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure Analysis of the IL-5 Ligand-Receptor Complex Reveals a Wrench-like Architecture for IL-5Ralpha.
Structure, 19, 2011
|
|
6IO9
 
 | | The structure of apo-UdgX | | Descriptor: | IRON/SULFUR CLUSTER, Phage SPO1 DNA polymerase-related protein, SULFATE ION | | Authors: | Xie, W, Tu, J. | | Deposit date: | 2018-10-29 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Suicide inactivation of the uracil DNA glycosylase UdgX by covalent complex formation.
Nat.Chem.Biol., 15, 2019
|
|
6IOB
 
 | | The structure of the H109A mutant of UdgX in complex with uracil | | Descriptor: | IRON/SULFUR CLUSTER, Phage SPO1 DNA polymerase-related protein, URACIL | | Authors: | Xie, W, Tu, J. | | Deposit date: | 2018-10-29 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Suicide inactivation of the uracil DNA glycosylase UdgX by covalent complex formation.
Nat.Chem.Biol., 15, 2019
|
|
1ECD
 
 | |
6IOD
 
 | | The structure of UdgX in complex with single-stranded DNA | | Descriptor: | DNA, IRON/SULFUR CLUSTER, Phage SPO1 DNA polymerase-related protein | | Authors: | Xie, W, Tu, J. | | Deposit date: | 2018-10-29 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Suicide inactivation of the uracil DNA glycosylase UdgX by covalent complex formation.
Nat.Chem.Biol., 15, 2019
|
|
3W28
 
 | | The high-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E251A mutant with xylotriose | | Descriptor: | Glycoside hydrolase family 10, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-xylopyranose | | Authors: | Han, X, Gao, J, Shang, N, Huang, C.-H, Ko, T.-P, Zhu, Z, Wiegel, J, Shao, W, Guo, R.-T. | | Deposit date: | 2012-11-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural and functional analyses of catalytic domain of GH10 xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Proteins, 81, 2013
|
|
1F5A
 
 | | CRYSTAL STRUCTURE OF MAMMALIAN POLY(A) POLYMERASE | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, POLY(A) POLYMERASE, ... | | Authors: | Martin, G, Keller, W, Doublie, S. | | Deposit date: | 2000-06-13 | | Release date: | 2000-09-13 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of mammalian poly(A) polymerase in complex with an analog of ATP.
EMBO J., 19, 2000
|
|
2M4J
 
 | | 40-residue beta-amyloid fibril derived from Alzheimer's disease brain | | Descriptor: | Amyloid beta A4 protein | | Authors: | Lu, J, Qiang, W, Meredith, S.C, Yau, W, Schweiters, C.D, Tycko, R. | | Deposit date: | 2013-02-05 | | Release date: | 2013-09-25 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Molecular Structure of beta-Amyloid Fibrils in Alzheimer's Disease Brain Tissue.
Cell(Cambridge,Mass.), 154, 2013
|
|
3W29
 
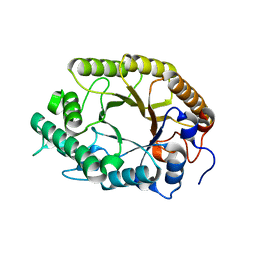 | | The high-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E251A mutant with xylotetraose | | Descriptor: | Glycoside hydrolase family 10, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-xylopyranose | | Authors: | Han, X, Gao, J, Shang, N, Huang, C.-H, Ko, T.-P, Zhu, Z, Wiegel, J, Shao, W, Guo, R.-T. | | Deposit date: | 2012-11-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural and functional analyses of catalytic domain of GH10 xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Proteins, 81, 2013
|
|
3W27
 
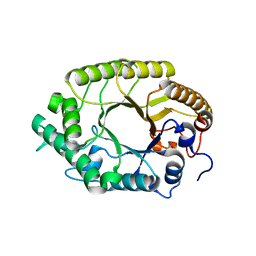 | | The high-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E251A mutant with xylobiose | | Descriptor: | Glycoside hydrolase family 10, beta-D-xylopyranose-(1-4)-alpha-D-xylopyranose | | Authors: | Han, X, Gao, J, Shang, N, Huang, C.-H, Ko, T.-P, Zhu, Z, Wiegel, J, Shao, W, Guo, R.-T. | | Deposit date: | 2012-11-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structural and functional analyses of catalytic domain of GH10 xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Proteins, 81, 2013
|
|
6AGK
 
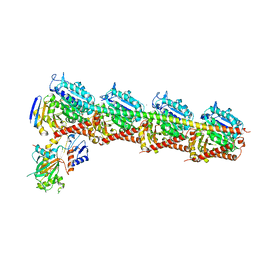 | | The structure of CH-II-77-tubulin complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Chen, H, Arnst, K, Wang, Y, Miller, D, Li, W. | | Deposit date: | 2018-08-13 | | Release date: | 2019-08-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Activity Relationship Study of Novel 6-Aryl-2-benzoyl-pyridines as Tubulin Polymerization Inhibitors with Potent Antiproliferative Properties.
J.Med.Chem., 63, 2020
|
|
3W25
 
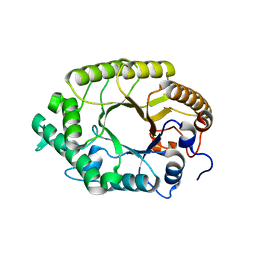 | | The high-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E146A mutant with xylobiose | | Descriptor: | Glycoside hydrolase family 10, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Han, X, Gao, J, Shang, N, Huang, C.-H, Ko, T.-P, Zhu, Z, Wiegel, J, Shao, W, Guo, R.-T. | | Deposit date: | 2012-11-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural and functional analyses of catalytic domain of GH10 xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Proteins, 81, 2013
|
|
3W26
 
 | | The high-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E146A mutant with xylotriose | | Descriptor: | Glycoside hydrolase family 10, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Han, X, Gao, J, Shang, N, Huang, C.-H, Ko, T.-P, Zhu, Z, Wiegel, J, Shao, W, Guo, R.-T. | | Deposit date: | 2012-11-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional analyses of catalytic domain of GH10 xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Proteins, 81, 2013
|
|
3W24
 
 | | The high-resolution crystal structure of TsXylA, intracellular xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485 | | Descriptor: | Glycoside hydrolase family 10 | | Authors: | Han, X, Gao, J, Shang, N, Huang, C.-H, Ko, T.-P, Zhu, Z, Wiegel, J, Shao, W, Guo, R.-T. | | Deposit date: | 2012-11-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural and functional analyses of catalytic domain of GH10 xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Proteins, 81, 2013
|
|
4L15
 
 | | Crystal structure of FIGL-1 AAA domain | | Descriptor: | Fidgetin-like protein 1, TRIS(HYDROXYETHYL)AMINOMETHANE | | Authors: | Peng, W, Lin, Z, Li, W, Lu, J, Shen, Y, Wang, C. | | Deposit date: | 2013-06-02 | | Release date: | 2013-09-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into the unusually strong ATPase activity of the AAA domain of the Caenorhabditis elegans fidgetin-like 1 (FIGL-1) protein.
J.Biol.Chem., 288, 2013
|
|
1ECO
 
 | |
1ECN
 
 | |
4QWP
 
 | | co-crystal structure of chitosanase OU01 with substrate | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Lyu, Q, Liu, W, Han, B. | | Deposit date: | 2014-07-17 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biochemical insights into the degradation mechanism of chitosan by chitosanase OU01.
Biochim.Biophys.Acta, 1850, 2015
|
|
4L16
 
 | | Crystal structure of FIGL-1 AAA domain in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Fidgetin-like protein 1 | | Authors: | Peng, W, Lin, Z, Li, W, Lu, J, Shen, Y, Wang, C. | | Deposit date: | 2013-06-02 | | Release date: | 2013-09-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the unusually strong ATPase activity of the AAA domain of the Caenorhabditis elegans fidgetin-like 1 (FIGL-1) protein.
J.Biol.Chem., 288, 2013
|
|
4L7J
 
 | | Diethylaminosulfur Trifluoride-Mediated Intramolecular Cyclization of 2-hydroxy-benzylureas to Fused Bicyclic Aminooxazoline Compounds and Evaluation of Their Biochemical Activity Against Beta-Secretase-1 (BACE-1) | | Descriptor: | 2-[(3aS,7aR)-2-amino-3a-(2,4-difluorophenyl)-3a,6,7,7a-tetrahydro[1,3]oxazolo[4,5-c]pyridin-5(4H)-yl]pyridine-3-carbonitrile, ACETATE ION, Beta-secretase 1 | | Authors: | Huestis, M.P, Liu, W, Volgraf, M, Purkey, H.E, Wang, W, Yu, C, Wu, P, Smith, D, Vigers, G, Dutcher, D, Geck Do, M.K, Hunt, K.W, Siu, M. | | Deposit date: | 2013-06-13 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Diethylaminosulfur Trifluoride-Mediated Intramolecular Cyclization of 2-hydroxycycloalkylureas to Fused Bicyclic Aminooxazoline Compounds and Evaluation of Their Biochemical Activity Against β-Secretase-1 (BACE-1)
Tetrahedron Lett., 2013
|
|
