2JCG
 
 | | Apo form of the catabolite control protein A (ccpA) from bacillus megaterium, with the DNA binding domain | | Descriptor: | CALCIUM ION, GLUCOSE-RESISTANCE AMYLASE REGULATOR | | Authors: | Singh, R.K, Panjikar, S, Palm, G.J, Hinrichs, W. | | Deposit date: | 2006-12-22 | | Release date: | 2007-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the Apo Form of the Catabolite Control Protein a (Ccpa) from Bacillus Megaterium with a DNA-Binding Domain.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
9AUJ
 
 | | Structure of SARS-CoV-2 Mpro mutant (S144A) in complex with Nirmatrelvir (PF-07321332) | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2024-02-29 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.486 Å) | | Cite: | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
9AUM
 
 | | Structure of SARS-CoV-2 Mpro mutant (T21I,S144A,T304I) in complex with Nirmatrelvir (PF-07321332) | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2024-02-29 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.539 Å) | | Cite: | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
9AUK
 
 | | Structure of SARS-CoV-2 Mpro mutant (A173V) in complex with Nirmatrelvir (PF-07321332) | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2024-02-29 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
9AUO
 
 | | Structure of SARS-CoV-2 Mpro mutant (L50F,T304I) | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2024-02-29 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.423 Å) | | Cite: | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
9AUN
 
 | | Structure of SARS-CoV-2 Mpro mutant (T21I,T304I) | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2024-02-29 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
8KBZ
 
 | |
8Y3U
 
 | |
121P
 
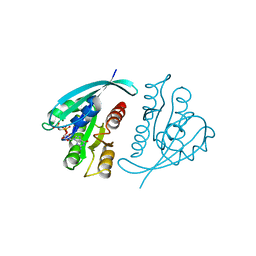 | | STRUKTUR UND GUANOSINTRIPHOSPHAT-HYDROLYSEMECHANISMUS DES C-TERMINAL VERKUERZTEN MENSCHLICHEN KREBSPROTEINS P21-H-RAS | | Descriptor: | H-RAS P21 PROTEIN, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Krengel, U, Scheffzek, K, Scherer, A, Kabsch, W, Wittinghofer, A, Pai, E.F. | | Deposit date: | 1991-06-06 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Struktur Und Guanosintriphosphat-Hydrolysemechanismus Des C-Terminal Verkuerzten Menschlichen Krebsproteins P21-H-Ras
Thesis, 1991
|
|
1AFR
 
 | |
4EL1
 
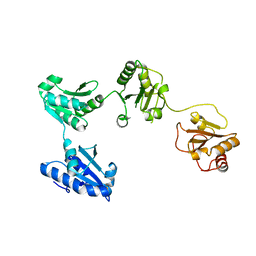 | | Crystal structure of oxidized hPDI (abb'xa') | | Descriptor: | Protein disulfide-isomerase | | Authors: | Wang, C, Li, W, Ren, J, Ke, H, Gong, W, Feng, W, Wang, C.-C. | | Deposit date: | 2012-04-10 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.883 Å) | | Cite: | Structural insights into the redox-regulated dynamic conformations of human protein disulfide isomerase
Antioxid Redox Signal, 19, 2013
|
|
4EKZ
 
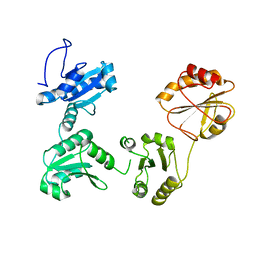 | | Crystal structure of reduced hPDI (abb'xa') | | Descriptor: | Protein disulfide-isomerase | | Authors: | Wang, C, Li, W, Ren, J, Ke, H, Gong, W, Feng, W, Wang, C.-C. | | Deposit date: | 2012-04-10 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural insights into the redox-regulated dynamic conformations of human protein disulfide isomerase
Antioxid Redox Signal, 19, 2013
|
|
1A3X
 
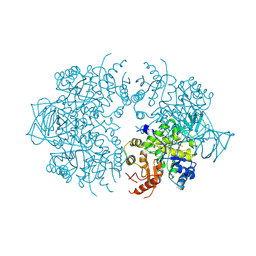 | | PYRUVATE KINASE FROM SACCHAROMYCES CEREVISIAE COMPLEXED WITH PG, MN2+ AND K+ | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, MANGANESE (II) ION, POTASSIUM ION, ... | | Authors: | Jurica, M.S, Mesecar, A, Heath, P.J, Shi, W, Nowak, T, Stoddard, B.L. | | Deposit date: | 1998-01-26 | | Release date: | 1998-05-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The allosteric regulation of pyruvate kinase by fructose-1,6-bisphosphate.
Structure, 6, 1998
|
|
7PQT
 
 | | Apo human Kv3.1 cryo-EM structure | | Descriptor: | POTASSIUM ION, Potassium voltage-gated channel subfamily C member 1 | | Authors: | Botte, M, Huber, S, Bucher, D, Klint, J.K, Rodriguez, D, Tagmose, L, Chami, M, Cheng, R, Hennig, M, Abdul Rhaman, W. | | Deposit date: | 2021-09-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Apo and ligand-bound high resolution Cryo-EM structures of the human Kv3.1 channel reveal a novel binding site for positive modulators.
Pnas Nexus, 1, 2022
|
|
7PQU
 
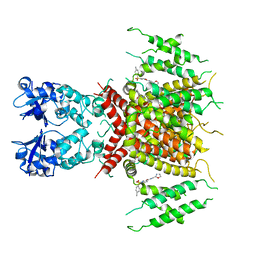 | | Ligand-bound human Kv3.1 cryo-EM structure (Lu AG00563) | | Descriptor: | 1-(4-methylphenyl)sulfonyl-N-(1,3-oxazol-2-ylmethyl)pyrrole-3-carboxamide, POTASSIUM ION, Potassium voltage-gated channel subfamily C member 1 | | Authors: | Botte, M, Huber, S, Bucher, D, Klint, J.K, Rodriguez, D, Tagmose, L, Chami, M, Cheng, R, Hennig, M, Abdul Rhaman, W. | | Deposit date: | 2021-09-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Apo and ligand-bound high resolution Cryo-EM structures of the human Kv3.1 channel reveal a novel binding site for positive modulators.
Pnas Nexus, 1, 2022
|
|
1AFV
 
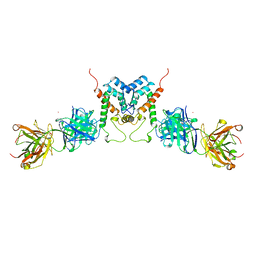 | | HIV-1 CAPSID PROTEIN (P24) COMPLEX WITH FAB25.3 | | Descriptor: | ANTIBODY FAB25.3 FRAGMENT (HEAVY CHAIN), ANTIBODY FAB25.3 FRAGMENT (LIGHT CHAIN), HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 CAPSID PROTEIN, ... | | Authors: | Momany, C, Kovari, L.C, Prongay, A.J, Keller, W, Gitti, R.K, Lee, B.M, Gorbalenya, A.E, Tong, L, Mcclure, J, Ehrlich, L.S, Summers, M.F, Carter, C, Rossmann, M.G. | | Deposit date: | 1997-03-14 | | Release date: | 1997-08-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal structure of dimeric HIV-1 capsid protein.
Nat.Struct.Biol., 3, 1996
|
|
1AH1
 
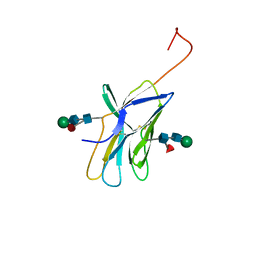 | | CTLA-4, NMR, 20 STRUCTURES | | Descriptor: | CTLA-4, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Metzler, W.J, Bajorath, J, Fenderson, W, Shaw, S.-Y, Peach, R, Constantine, K.L, Naemura, J, Leytze, G, Lavoie, T.B, Mueller, L, Linsley, P.S. | | Deposit date: | 1997-04-11 | | Release date: | 1998-04-15 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human CTLA-4 and delineation of a CD80/CD86 binding site conserved in CD28.
Nat.Struct.Biol., 4, 1997
|
|
1A6S
 
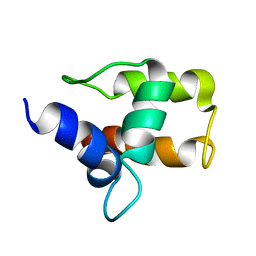 | | M-DOMAIN FROM GAG POLYPROTEIN OF ROUS SARCOMA VIRUS, NMR, 20 STRUCTURES | | Descriptor: | GAG POLYPROTEIN | | Authors: | Mcdonnell, J.M, Fushman, D, Cahill, S.M, Zhou, W, Wolven, A, Wilson, C.B, Nelle, T.D, Resh, M.D, Wills, J, Cowburn, D. | | Deposit date: | 1998-03-02 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the bioactive retroviral M domain from Rous sarcoma virus
J.Mol.Biol., 279, 1998
|
|
6MPV
 
 | |
1A69
 
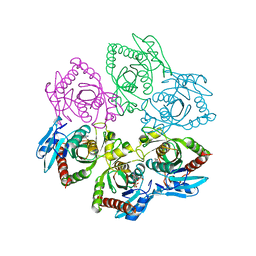 | | PURINE NUCLEOSIDE PHOSPHORYLASE IN COMPLEX WITH FORMYCIN B AND SULPHATE (PHOSPHATE) | | Descriptor: | FORMYCIN B, PURINE NUCLEOSIDE PHOSPHORYLASE, SULFATE ION | | Authors: | Koellner, G, Luic, M, Shugar, D, Saenger, W, Bzowska, A. | | Deposit date: | 1998-03-08 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the ternary complex of E. coli purine nucleoside phosphorylase with formycin B, a structural analogue of the substrate inosine, and phosphate (Sulphate) at 2.1 A resolution.
J.Mol.Biol., 280, 1998
|
|
1AHJ
 
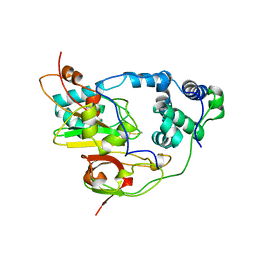 | | NITRILE HYDRATASE | | Descriptor: | FE (III) ION, NITRILE HYDRATASE (SUBUNIT ALPHA), NITRILE HYDRATASE (SUBUNIT BETA) | | Authors: | Huang, W, Schneider, G, Lindqvist, Y. | | Deposit date: | 1997-04-05 | | Release date: | 1998-04-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of nitrile hydratase reveals a novel iron centre in a novel fold.
Structure, 5, 1997
|
|
1A4T
 
 | | SOLUTION STRUCTURE OF PHAGE P22 N PEPTIDE-BOX B RNA COMPLEX, NMR, 20 STRUCTURES | | Descriptor: | 20-MER BASIC PEPTIDE, BOXB RNA | | Authors: | Cai, Z, Gorin, A.A, Frederick, R, Ye, X, Hu, W, Majumdar, A, Kettani, A, Patel, D.J. | | Deposit date: | 1998-02-04 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of P22 transcriptional antitermination N peptide-boxB RNA complex.
Nat.Struct.Biol., 5, 1998
|
|
1A6R
 
 | | GAL6 (YEAST BLEOMYCIN HYDROLASE) MUTANT C73A | | Descriptor: | GAL6, SULFATE ION | | Authors: | Joshua-Tor, L, Zheng, W, Johnston, S.A. | | Deposit date: | 1998-02-27 | | Release date: | 1998-10-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The unusual active site of Gal6/bleomycin hydrolase can act as a carboxypeptidase, aminopeptidase, and peptide ligase.
Cell(Cambridge,Mass.), 93, 1998
|
|
1AJ4
 
 | | STRUCTURE OF CALCIUM-SATURATED CARDIAC TROPONIN C, NMR, 1 STRUCTURE | | Descriptor: | CALCIUM ION, TROPONIN C | | Authors: | Sia, S.K, Li, M.X, Spyracopoulos, L, Gagne, S.M, Liu, W, Putkey, J.A, Sykes, B.D. | | Deposit date: | 1997-05-14 | | Release date: | 1998-05-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of cardiac muscle troponin C unexpectedly reveals a closed regulatory domain.
J.Biol.Chem., 272, 1997
|
|
1B6Z
 
 | | 6-PYRUVOYL TETRAHYDROPTERIN SYNTHASE | | Descriptor: | 6-pyruvoyl tetrahydropterin synthase, ZINC ION | | Authors: | Ploom, T, Thoeny, B, Yim, J, Lee, S, Nar, H, Leimbacher, W, Huber, R, Richardson, J, Auerbach, G. | | Deposit date: | 1999-01-18 | | Release date: | 2000-01-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic and kinetic investigations on the mechanism of 6-pyruvoyl tetrahydropterin synthase.
J.Mol.Biol., 286, 1999
|
|
