3TEP
 
 | | LytR-CPS2a-Psr family protein with bound octaprenyl pyrophosphate lipid and magnesium ion | | Descriptor: | (2Z,6Z,10Z,14Z,18Z,22Z,26Z)-3,7,11,15,19,23,27,31-octamethyldotriaconta-2,6,10,14,18,22,26,30-octaen-1-yl trihydrogen diphosphate, 1,2-ETHANEDIOL, CPS2A, ... | | Authors: | Kawai, Y, Marles-Wright, J, Cleverley, R.M, Emmins, R, Ishikawa, S, Kuwano, M, Heinz, N, Khai Bui, N, Hoyland, C.N, Ogasawara, N, Lewis, R.J, Vollmer, W, Daniel, R.A, Errington, J. | | Deposit date: | 2011-08-15 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A widespread family of bacterial cell wall assembly proteins.
Embo J., 30, 2011
|
|
3SST
 
 | |
3SRY
 
 | |
3SVE
 
 | |
3SVB
 
 | |
3SZU
 
 | | IspH:HMBPP complex structure of E126Q mutant | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, FE3-S4 CLUSTER | | Authors: | Span, I, Graewert, T, Bacher, A, Eisenreich, W, Groll, M. | | Deposit date: | 2011-07-19 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structures of Mutant IspH Proteins Reveal a Rotation of the Substrate's Hydroxymethyl Group during Catalysis.
J.Mol.Biol., 416, 2012
|
|
3SZG
 
 | | Crystal structure of C176A glutamine-dependent NAD+ synthetase from M. tuberculosis bound to AMP/PPi and NaAD+ | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, Glutamine-dependent NAD(+) synthetase, ... | | Authors: | Chuenchor, W, Doukov, T, Gerratana, B. | | Deposit date: | 2011-07-19 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Regulation of the intersubunit ammonia tunnel in Mycobacterium tuberculosis glutamine-dependent NAD+ synthetase.
Biochem.J., 443, 2012
|
|
3SOQ
 
 | | The structure of the first YWTD beta propeller domain of LRP6 in complex with a DKK1 peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Dickkopf-related protein 1, ... | | Authors: | Wang, W, Bourhis, E, Zhang, Y, Rouge, L, Wu, Y, Franke, Y, Cochran, A.G. | | Deposit date: | 2011-06-30 | | Release date: | 2011-09-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Wnt antagonists bind through a short peptide to the first beta-propeller domain of LRP5/6.
Structure, 19, 2011
|
|
3SV5
 
 | | Engineered medium-affinity halide-binding protein derived from YFP: iodide complex | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Green fluorescent protein, ... | | Authors: | Wang, W, Grimley, J.S, Beese, L.S, Hellinga, H.W. | | Deposit date: | 2011-07-12 | | Release date: | 2012-07-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Visualization of Synaptic Inhibition with an Optogenetic Sensor Developed by Cell-Free Protein Engineering Automation.
J.Neurosci., 33, 2013
|
|
3TH8
 
 | | Structure of E. coli undecaprenyl diphosphate synthase complexed with BPH-1063 | | Descriptor: | (2Z)-4-({3-[3-(hexyloxy)phenyl]propyl}amino)-2-hydroxy-4-oxobut-2-enoic acid, Undecaprenyl pyrophosphate synthase | | Authors: | Cao, R, Zhu, W, Zhang, Y, Oldfield, E. | | Deposit date: | 2011-08-18 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.114 Å) | | Cite: | HIV-1 Integrase Inhibitor-Inspired Antibacterials Targeting Isoprenoid Biosynthesis.
ACS Med Chem Lett, 3, 2012
|
|
3TAN
 
 | |
3TEL
 
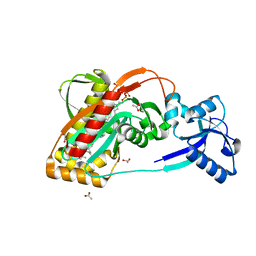 | | LytR-CPS2A-Psr family protein with bound octaprenyl pyrophosphate lipid and manganese ion | | Descriptor: | (2Z,6Z,10Z,14Z,18Z,22Z,26Z)-3,7,11,15,19,23,27,31-octamethyldotriaconta-2,6,10,14,18,22,26,30-octaen-1-yl trihydrogen diphosphate, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Kawai, Y, Marles-Wright, J, Cleverley, R.M, Emmins, R, Ishikawa, S, Kuwano, M, Heinz, N, Khai Bui, N, Hoyland, C.N, Ogasawara, N, Lewis, R.J, Vollmer, W, Daniel, R.A, Errington, J. | | Deposit date: | 2011-08-15 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A widespread family of bacterial cell wall assembly proteins.
Embo J., 30, 2011
|
|
3TET
 
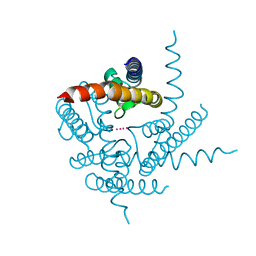 | | Crystal Structure of NaK2K Channel Y66F Mutant | | Descriptor: | POTASSIUM ION, Potassium channel protein | | Authors: | Sauer, D.B, Zeng, W, Raghunathan, S, Jiang, Y. | | Deposit date: | 2011-08-15 | | Release date: | 2011-10-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein interactions central to stabilizing the K+ channel selectivity filter in a four-sited configuration for selective K+ permeation.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3THX
 
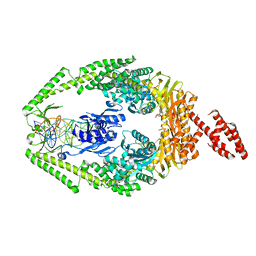 | | Human MutSbeta complexed with an IDL of 3 bases (Loop3) and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA Loop3 minus strand, DNA Loop3 plus strand, ... | | Authors: | Yang, W. | | Deposit date: | 2011-08-19 | | Release date: | 2011-12-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanism of mismatch recognition revealed by human MutSbeta bound to unpaired DNA loops
Nat.Struct.Mol.Biol., 19, 2012
|
|
3THV
 
 | |
3TJT
 
 | | Crystal Structure Analysis of the superoxide dismutase from Clostridium difficile | | Descriptor: | FE (III) ION, Superoxide dismutase | | Authors: | Li, W, Lei, C, Ying, T.L, Wang, H.F, Tan, X.S. | | Deposit date: | 2011-08-25 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Crystal Structure of the superoxide dismutase from Clostridium difficile
To be Published
|
|
3TKL
 
 | | Crystal structure of the GTP-bound Rab1a in complex with the coiled-coil domain of LidA from Legionella pneumophila | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, LidA protein, substrate of the Dot/Icm system, ... | | Authors: | Cheng, W, Yin, K, Lu, D, Li, B, Zhu, D, Chen, Y, Zhang, H, Xu, S, Chai, J, Gu, L. | | Deposit date: | 2011-08-27 | | Release date: | 2012-06-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.183 Å) | | Cite: | Structural insights into a unique Legionella pneumophila effector LidA recognizing both GDP and GTP bound Rab1 in their active state
Plos Pathog., 8, 2012
|
|
3SSK
 
 | |
3SS0
 
 | |
3SSH
 
 | |
3SVC
 
 | | Engineered medium-affinity halide-binding protein derived from YFP: chloride complex | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Green fluorescent protein | | Authors: | Wang, W, Grimley, J.S, Beese, L.S, Hellinga, H.W. | | Deposit date: | 2011-07-12 | | Release date: | 2012-07-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Determination of engineered chloride-binding site structures in fluorescent proteins reveals principles of halide recognition
To be Published
|
|
3SSL
 
 | |
3SSY
 
 | |
3Q7J
 
 | | Engineered Thermoplasma Acidophilum F3 factor mimics human aminopeptidase N (APN) as a target for anticancer drug development | | Descriptor: | L-phenylalanyl-N6-[(benzyloxy)carbonyl]-N1-hydroxy-L-lysinamide, Tricorn protease-interacting factor F3, ZINC ION | | Authors: | Su, J, Wang, Q, Feng, J, Zhang, C, Zhu, D, We, T, Xu, W, Gu, L. | | Deposit date: | 2011-01-05 | | Release date: | 2011-07-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Engineered Thermoplasma acidophilum factor F3 mimics human aminopeptidase N (APN) as a target for anticancer drug development
Bioorg.Med.Chem., 19, 2011
|
|
3QTD
 
 | | Crystal structure of putative modulator of gyrase (PmbA) from Pseudomonas aeruginosa PAO1 | | Descriptor: | GLYCEROL, PmbA protein | | Authors: | Tkaczuk, K.L, Chruszcz, M, Evdokimova, E, Liu, F, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-22 | | Release date: | 2011-03-30 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of putative modulator of gyrase (PmbA) from Pseudomonas aeruginosa PAO1
To be Published
|
|
