3JUI
 
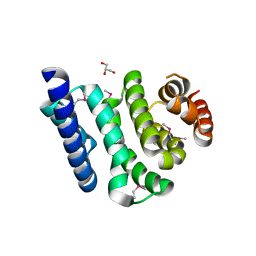 | | Crystal Structure of the C-terminal Domain of Human Translation Initiation Factor eIF2B epsilon Subunit | | Descriptor: | GLYCEROL, Translation initiation factor eIF-2B subunit epsilon | | Authors: | Wei, J, Xu, H, Zhang, C, Wang, M, Gao, F, Gong, W. | | Deposit date: | 2009-09-15 | | Release date: | 2009-10-06 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the C-terminal Domain of Human Translation Initiation Factor eIF2B epsilon Subunit
To be published
|
|
8V6Q
 
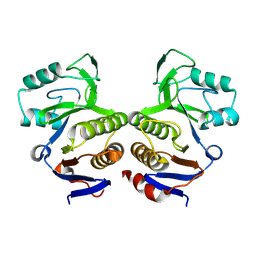 | | Crystal structure of EcThsA in ligand-free state | | Descriptor: | Thoeris protein ThsA Macro domain-containing protein | | Authors: | Shi, Y, Masic, V, Mosaiab, T, Goulart, C.C, Hartley-Tassell, L, Sorbello, M, Vasquez, V, Mishra, B.P, Holt, S, Gu, W, Kobe, B, Ve, T. | | Deposit date: | 2023-12-02 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural characterization of macro domain-containing Thoeris antiphage defense systems.
Sci Adv, 10, 2024
|
|
8V6S
 
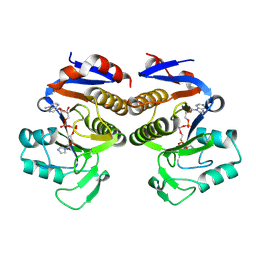 | | Crystal structure of PcThsA in complex with Imidazole Adenine Dinucleotide | | Descriptor: | Thoeris protein ThsA Macro domain-containing protein, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-imidazol-1-yl-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Shi, Y, Masic, V, Mosaiab, T, Goulart, C.C, Hartley-Tassell, L, Sorbello, M, Vasquez, E, Mishra, B.P, Holt, S, Gu, W, Kobe, B, Ve, T. | | Deposit date: | 2023-12-02 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural characterization of macro domain-containing Thoeris antiphage defense systems.
Sci Adv, 10, 2024
|
|
4QAU
 
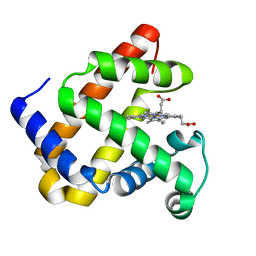 | | Crystal structure of F43Y mutant of sperm whale myoglobin | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lin, Y, Tan, X, Li, W. | | Deposit date: | 2014-05-06 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.597 Å) | | Cite: | A Novel Tyrosine-Heme C-O Covalent Linkage in F43Y Myoglobin: A New Post-translational Modification of Heme Proteins
Chembiochem, 16, 2015
|
|
8V6T
 
 | | Crystal structure of EcThsB2 | | Descriptor: | BROMIDE ION, Molecular chaperone Tir | | Authors: | Shi, Y, Masic, V, Mosaiab, T, Goulart, C.C, Hartley-Tassell, L, Sorbello, M, Vasquez, E, Mishra, B.P, Holt, S, Gu, W, Kobe, B, Ve, T. | | Deposit date: | 2023-12-02 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural characterization of macro domain-containing Thoeris antiphage defense systems.
Sci Adv, 10, 2024
|
|
8V6R
 
 | | Crystal structure of EcThsA in complex with ADPR | | Descriptor: | Thoeris protein ThsA Macro domain-containing protein, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Shi, Y, Masic, V, Mosaiab, T, Goulart, C.C, Hartley-Tassell, L, Sorbello, M, Vasquez, E, Mishra, B.P, Holt, S, Gu, W, Kobe, B, Ve, T. | | Deposit date: | 2023-12-02 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural characterization of macro domain-containing Thoeris antiphage defense systems.
Sci Adv, 10, 2024
|
|
3KJF
 
 | | Caspase 3 Bound to a covalent inhibitor | | Descriptor: | (3S)-3-({[(5S,10aS)-2-{(2S)-4-carboxy-2-[(phenylacetyl)amino]butyl}-1,3-dioxo-2,3,5,7,8,9,10,10a-octahydro-1H-[1,2,4]triazolo[1,2-a]cinnolin-5-yl]carbonyl}amino)-4-oxopentanoic acid, Caspase-3 | | Authors: | Kamtekar, S, Watt, W, Finzel, B.C, Harris, M.S, Blinn, J, Wang, Z, Tomasselli, A.G. | | Deposit date: | 2009-11-03 | | Release date: | 2010-08-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic and structural characterization of caspase-3 and caspase-8 inhibition by a novel class of irreversible inhibitors.
Biochim.Biophys.Acta, 1804, 2010
|
|
3KJN
 
 | | Caspase 8 bound to a covalent inhibitor | | Descriptor: | (3S)-3-({[(5S)-2-{2-[(1H-benzimidazol-5-ylcarbonyl)amino]ethyl}-7-(cyclohexylmethyl)-1,3-dioxo-2,3,5,8-tetrahydro-1H-[1,2,4]triazolo[1,2-a]pyridazin-5-yl]carbonyl}amino)-4-oxopentanoic acid, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Caspase-8 | | Authors: | Kamtekar, S, Watt, W, Finzel, B.C, Harris, M.S, Blinn, J, Wang, Z, Tomasselli, A.G. | | Deposit date: | 2009-11-03 | | Release date: | 2010-08-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Kinetic and structural characterization of caspase-3 and caspase-8 inhibition by a novel class of irreversible inhibitors.
Biochim.Biophys.Acta, 1804, 2010
|
|
3KH4
 
 | |
3KLK
 
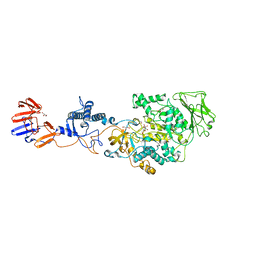 | | Crystal structure of Lactobacillus reuteri N-terminally truncated glucansucrase GTF180 in triclinic apo- form | | Descriptor: | CALCIUM ION, GLYCEROL, Glucansucrase | | Authors: | Vujicic-Zagar, A, Pijning, T, Kralj, S, Eeuwema, W, Dijkhuizen, L, Dijkstra, B.W. | | Deposit date: | 2009-11-08 | | Release date: | 2010-11-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of a 117 kDa glucansucrase fragment provides insight into evolution and product specificity of GH70 enzymes
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3KBH
 
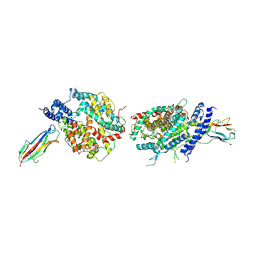 | | Crystal structure of NL63 respiratory coronavirus receptor-binding domain complexed with its human receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Wu, K, Li, W, Peng, G, Li, F. | | Deposit date: | 2009-10-20 | | Release date: | 2009-12-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Crystal structure of NL63 respiratory coronavirus receptor-binding domain complexed with its human receptor.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3KMR
 
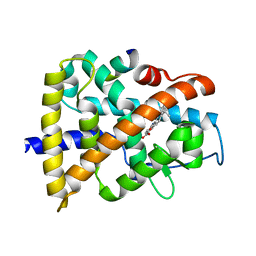 | | Crystal structure of RARalpha ligand binding domain in complex with an agonist ligand (Am580) and a coactivator fragment | | Descriptor: | 4-{[(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)carbonyl]amino}benzoic acid, Nuclear receptor coactivator 1, Retinoic acid receptor alpha | | Authors: | Bourguet, W, Teyssier, C. | | Deposit date: | 2009-11-11 | | Release date: | 2010-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A unique secondary-structure switch controls constitutive gene repression by retinoic acid receptor.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KEF
 
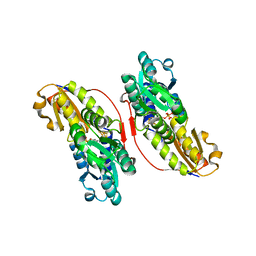 | | Crystal structure of IspH:DMAPP-complex | | Descriptor: | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, DIMETHYLALLYL DIPHOSPHATE, FE3-S4 CLUSTER | | Authors: | Groll, M, Graewert, T, Span, I, Eisenreich, W, Bacher, A. | | Deposit date: | 2009-10-26 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Probing the reaction mechanism of IspH protein by x-ray structure analysis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
8U45
 
 | | Crystal Structure Analysis of Aspergillus fumigatus alkaline protease | | Descriptor: | Alkaline protease 1, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Fernandez, D, Diec, D.D.L, Guo, W, Russi, S. | | Deposit date: | 2023-09-08 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Targeting Aspergillus allergen oryzin with a chemical probe at atomic precision.
Sci Rep, 13, 2023
|
|
4Q96
 
 | | CID of human RPRD1B in complex with an unmodified CTD peptide | | Descriptor: | RPB1-CTD, Regulation of nuclear pre-mRNA domain-containing protein 1B, SULFATE ION, ... | | Authors: | Ni, Z, Xu, C, Tempel, W, El Bakkouri, M, Loppnau, P, Guo, X, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Greenblatt, J.F, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-04-29 | | Release date: | 2014-06-04 | | Last modified: | 2014-08-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | RPRD1A and RPRD1B are human RNA polymerase II C-terminal domain scaffolds for Ser5 dephosphorylation.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4PW8
 
 | | Human tryptophan 2,3-dioxygenase | | Descriptor: | COBALT (II) ION, Tryptophan 2,3-dioxygenase | | Authors: | Meng, B, Wu, D, Gu, J.H, Ouyang, S.Y, Ding, W, Liu, Z.J. | | Deposit date: | 2014-03-19 | | Release date: | 2014-08-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Structural and functional analyses of human tryptophan 2,3-dioxygenase
Proteins, 82, 2014
|
|
3KEM
 
 | | Crystal structure of IspH:IPP complex | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, FE3-S4 CLUSTER | | Authors: | Groll, M, Graewert, T, Span, I, Eisenreich, W, Bacher, A. | | Deposit date: | 2009-10-26 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the reaction mechanism of IspH protein by x-ray structure analysis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3KE9
 
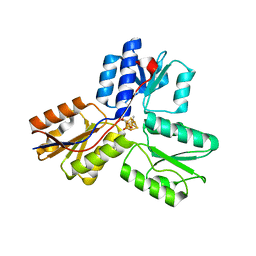 | | Crystal structure of IspH:Intermediate-complex | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IRON/SULFUR CLUSTER | | Authors: | Groll, M, Graewert, T, Span, I, Eisenreich, W, Bacher, A. | | Deposit date: | 2009-10-24 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the reaction mechanism of IspH protein by x-ray structure analysis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3KEL
 
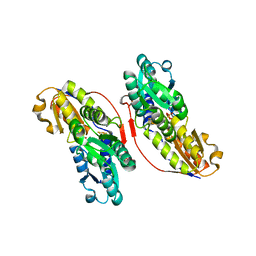 | | Crystal Structure of IspH:PP complex | | Descriptor: | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, FE3-S4 CLUSTER, PYROPHOSPHATE 2- | | Authors: | Groll, M, Graewert, T, Span, I, Eisenreich, W, Bacher, A. | | Deposit date: | 2009-10-26 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Probing the reaction mechanism of IspH protein by x-ray structure analysis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
8U5G
 
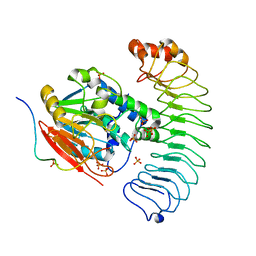 | | Crystal structure of the co-expressed SDS22:PP1:I3 complex | | Descriptor: | E3 ubiquitin-protein ligase PPP1R11, FE (III) ION, PHOSPHATE ION, ... | | Authors: | Choy, M.S, Peti, W, Page, R. | | Deposit date: | 2023-09-12 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The SDS22:PP1:I3 complex: SDS22 binding to PP1 loosens the active site metal to prime metal exchange.
J.Biol.Chem., 300, 2023
|
|
4QE9
 
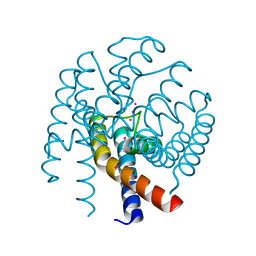 | | Open MthK pore structure soaked in 10 mM Ba2+/100 mM K+ | | Descriptor: | Calcium-gated potassium channel MthK, POTASSIUM ION | | Authors: | Guo, R, Zeng, W, Cui, H, Chen, L, Ye, S. | | Deposit date: | 2014-05-15 | | Release date: | 2014-07-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Ionic interactions of Ba2+ blockades in the MthK K+ channel
J.Gen.Physiol., 144, 2014
|
|
4QGM
 
 | | Acireductone dioxygenase from Bacillus anthracis with cadmium ion in active center | | Descriptor: | Acireductone dioxygenase, CADMIUM ION | | Authors: | Milaczewska, A.M, Chruszcz, M, Shabalin, I.G, Cooper, D.R, Borowski, T, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-05-23 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Acireductone dioxygenase from Bacillus anthracis with cadmium ion in active center
To be Published
|
|
3J7M
 
 | | Virus model of brome mosaic virus (first half data set) | | Descriptor: | Capsid protein | | Authors: | Wang, Z, Hryc, C, Bammes, B, Afonine, P.V, Jakana, J, Chen, D.H, Liu, X, Baker, M.L, Kao, C, Ludtke, S.J, Schmid, M.F, Adams, P.D, Chiu, W. | | Deposit date: | 2014-07-18 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | An atomic model of brome mosaic virus using direct electron detection and real-space optimization.
Nat Commun, 5, 2014
|
|
7W1N
 
 | | Complex structure of a leaf-branch compost cutinase variant LCC ICCG_KRP | | Descriptor: | 1,2-ETHANEDIOL, BICINE, Leaf-branch compost cutinase | | Authors: | Niu, D, Zeng, W, Huang, J.W, Chen, C.C, Liu, W.D, Guo, R.T. | | Deposit date: | 2021-11-19 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Substrate-Binding Mode of a Thermophilic PET Hydrolase and Engineering the Enzyme to Enhance the Hydrolytic Efficacy.
Acs Catalysis, 12, 2022
|
|
7WI6
 
 | | Cryo-EM structure of LY341495/NAM-bound mGlu3 | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 3 | | Authors: | Fang, W, Yang, F, Xu, C.J, Ling, S.L, Lin, L, Zhou, Y.X, Sun, W.J, Wang, X.M, Liu, P, Rondard, P, Pan, S, Pin, J.P, Tian, C.L, Liu, J.F. | | Deposit date: | 2022-01-03 | | Release date: | 2022-03-16 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structural basis of the activation of metabotropic glutamate receptor 3.
Cell Res., 32, 2022
|
|
