7XK2
 
 | | Cryo-EM Structure of Human Niacin Receptor HCA2-Gi protein complex | | Descriptor: | 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yang, Y, Kang, H.J, Gao, R.G, Wang, J.J, Han, G.W, DiBerto, J.F, Wu, L.J, Tong, J.H, Qu, L, Wu, Y.R, Pileski, R, Li, X.M, Zhang, X.C, Zhao, S.W, Kenakin, T, Wang, Q, Stevens, R.C, Peng, W, Roth, B.L, Rao, Z.H, Liu, Z.J. | | Deposit date: | 2022-04-19 | | Release date: | 2023-02-22 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the human niacin receptor HCA2-G i signalling complex.
Nat Commun, 14, 2023
|
|
8J0H
 
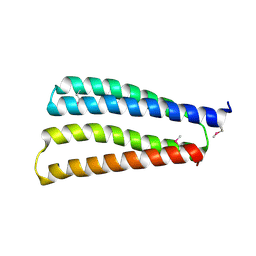 | |
7X3O
 
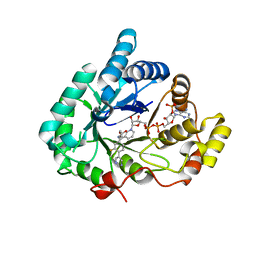 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound S07054 | | Descriptor: | (2~{R})-2-(3-fluoranyl-4-pyrimidin-5-yl-phenyl)butanoic acid, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Fang, P, Sun, H. | | Deposit date: | 2022-03-01 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal structure of Aldo-keto reductase 1C3 complexed with compound S07054
To Be Published
|
|
5BQT
 
 | | Structure of TrmBL2, an archaeal chromatin protein, shows a novel mode of DNA binding. | | Descriptor: | CALCIUM ION, Putative HTH-type transcriptional regulator TrmBL2 | | Authors: | Ahmad, M.U, Diederichs, K, Welte, W. | | Deposit date: | 2015-05-29 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Insights into Nonspecific Binding of DNA by TrmBL2, an Archaeal Chromatin Protein.
J.Mol.Biol., 427, 2015
|
|
7X3L
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound S07044 | | Descriptor: | (2~{R})-2-[4-(3-fluoranyl-4-methyl-phenyl)-3-(trifluoromethyl)phenyl]butanoic acid, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Fang, P, Sun, H. | | Deposit date: | 2022-03-01 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Development of Biaryl-Containing Aldo-Keto Reductase 1C3 (AKR1C3) Inhibitors for Reversing AKR1C3-Mediated Drug Resistance in Cancer Treatment.
J.Med.Chem., 66, 2023
|
|
7X3M
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound S07045 | | Descriptor: | (2~{R})-2-[4-[3,5-bis(chloranyl)phenyl]-3-(trifluoromethyl)phenyl]butanoic acid, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Fang, P, Sun, H. | | Deposit date: | 2022-03-01 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | Development of Biaryl-Containing Aldo-Keto Reductase 1C3 (AKR1C3) Inhibitors for Reversing AKR1C3-Mediated Drug Resistance in Cancer Treatment.
J.Med.Chem., 66, 2023
|
|
7X5P
 
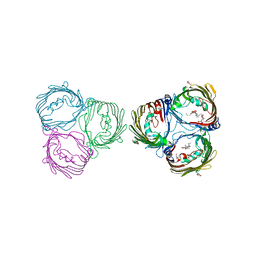 | | Truncated VhChiP (1-19aa) in complex with doxycycline | | Descriptor: | (4S,4AR,5S,5AR,6R,12AS)-4-(DIMETHYLAMINO)-3,5,10,12,12A-PENTAHYDROXY-6-METHYL-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2-CARBOXAMIDE, (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Chitoporin, ... | | Authors: | Sanram, S, Robinson, C.R, Aunkham, A, Suginta, W. | | Deposit date: | 2022-03-05 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure and function of truncated VhChiP
To Be Published
|
|
7X5Q
 
 | | Apo Truncated VhChiP (Delta 1-19) in complex with peptide (DGANSDAAK) | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, ASP-GLY-ALA-ASN-SER-ASP, Chitoporin, ... | | Authors: | Sanram, S, Aunkham, A, Suginta, W, Robinson, R.C. | | Deposit date: | 2022-03-05 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and function of truncated VhChiP
To Be Published
|
|
7XFV
 
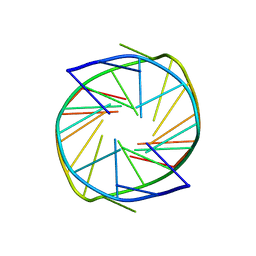 | |
8QXS
 
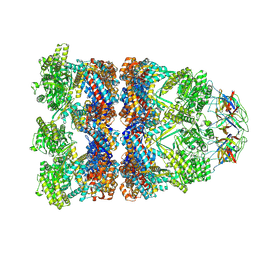 | | CryoEM structure of a GroEL14-GroES7 complex in presence of ADP-BeFx with wide GroEL7 trans ring conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chaperonin GroEL, ... | | Authors: | Wagner, J, Caravajal, A.I, Beck, F, Bracher, A, Wan, W, Bohn, S, Koerner, R, Baumeister, W, Fernandez-Busnadiego, R, Hartl, F.U. | | Deposit date: | 2023-10-25 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Visualizing chaperonin function in situ by cryo-electron tomography.
Nature, 633, 2024
|
|
8QXT
 
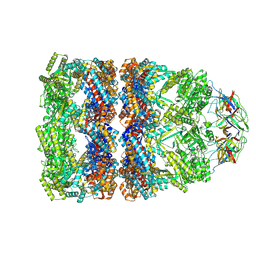 | | CryoEM structure of a GroEL14-GroES7 complex in presence of ADP-BeFx with narrow GroEL7 trans ring conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chaperonin GroEL, ... | | Authors: | Wagner, J, Caravajal, A.I, Beck, F, Bracher, A, Wan, W, Bohn, S, Koerner, R, Baumeister, W, Fernandez-Busnadiego, R, Hartl, F.U. | | Deposit date: | 2023-10-25 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Visualizing chaperonin function in situ by cryo-electron tomography.
Nature, 633, 2024
|
|
7X8U
 
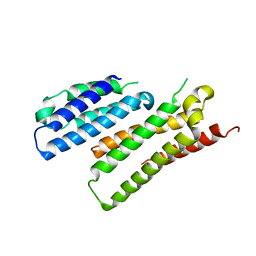 | |
8QXU
 
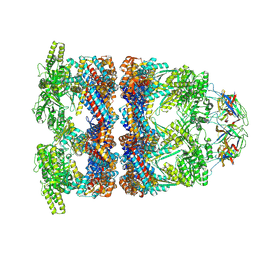 | | In situ structure average of GroEL14-GroES7 complexes with wide GroEL7 trans ring conformation in Escherichia coli cytosol obtained by cryo electron tomography | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Chaperonin GroEL, ... | | Authors: | Wagner, J, Caravajal, A.I, Beck, F, Bracher, A, Wan, W, Bohn, S, Koerner, R, Baumeister, W, Fernandez-Busnadiego, R, Hartl, F.U. | | Deposit date: | 2023-10-25 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Visualizing chaperonin function in situ by cryo-electron tomography.
Nature, 633, 2024
|
|
8QXV
 
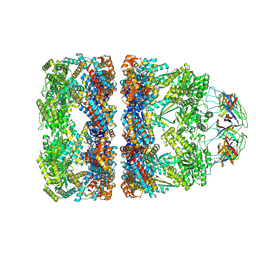 | | In situ structure average of GroEL14-GroES7 complexes with narrow GroEL7 trans ring conformation in Escherichia coli cytosol obtained by cryo electron tomography | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Chaperonin GroEL, ... | | Authors: | Wagner, J, Caravajal, A.I, Beck, F, Bracher, A, Wan, W, Bohn, S, Koerner, R, Baumeister, W, Fernandez-Busnadiego, R, Hartl, F.U. | | Deposit date: | 2023-10-25 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (13.6 Å) | | Cite: | Visualizing chaperonin function in situ by cryo-electron tomography.
Nature, 633, 2024
|
|
5BOX
 
 | | Structure of TrmBL2, an archaeal chromatin protein, shows a novel mode of DNA binding. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (25-MER), DNA TGM (25-MER), ... | | Authors: | Ahmad, M.U, Diederichs, K, Welte, W. | | Deposit date: | 2015-05-27 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights into Nonspecific Binding of DNA by TrmBL2, an Archaeal Chromatin Protein.
J.Mol.Biol., 427, 2015
|
|
7XLC
 
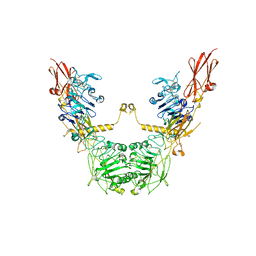 | | conformation II of apo-IGF1R states | | Descriptor: | Insulin-like growth factor 1 receptor | | Authors: | Xi, Z, Cang, W. | | Deposit date: | 2022-04-21 | | Release date: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | conformation II of apo-IGF1R states
To Be Published
|
|
7XL6
 
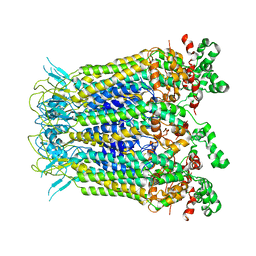 | |
7XJJ
 
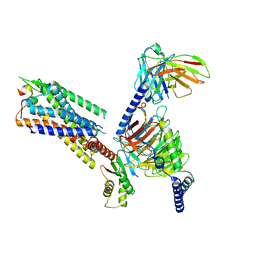 | | Cryo-EM structure of the galanin-bound GALR1-miniGo complex | | Descriptor: | G protein subunit alpha o1,Guanine nucleotide-binding protein G(o) subunit alpha, Galanin, Galanin receptor type 1, ... | | Authors: | Jiang, W, Zheng, S. | | Deposit date: | 2022-04-18 | | Release date: | 2023-05-03 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into galanin receptor signaling.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XMN
 
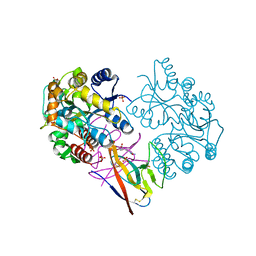 | | Structure of SARS-CoV-2 ORF8 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Maltodextrin-binding protein, ... | | Authors: | Chen, X, Xu, W. | | Deposit date: | 2022-04-26 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Glycosylated, Lipid-Binding, CDR-Like Domains of SARS-CoV-2 ORF8 Indicate Unique Sites of Immune Regulation.
Microbiol Spectr, 11, 2023
|
|
7XP9
 
 | | Phytophthora infesfans RxLR effector AVRvnt1 | | Descriptor: | RxLR effector protein Avr-vnt11 | | Authors: | Xing, W, Hu, Q, Zhou, J, Yao, D. | | Deposit date: | 2022-05-04 | | Release date: | 2023-06-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Chloroplast Protein GLYK Hijacked by Phytophthora Infestans Effector AVRvnt1 in Cytoplasm to Activate NLR
To Be Published
|
|
5BMS
 
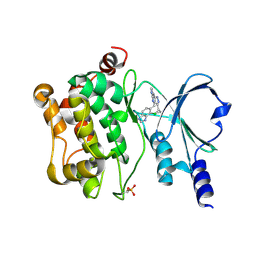 | |
7X2Z
 
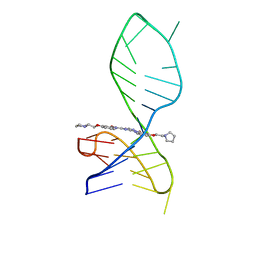 | | NMR solution structure of the 1:1 complex of a pyridostatin derivative (PyPDS) bound to a G-quadruplex MYT1L | | Descriptor: | 4-(2-azanylethoxy)-N2,N6-bis[4-(2-pyrrolidin-1-ylethoxy)quinolin-2-yl]pyridine-2,6-dicarboxamide, G-quadruplex DNA MYT1L | | Authors: | Liu, L.-Y, Mao, Z.-W, Liu, W. | | Deposit date: | 2022-02-26 | | Release date: | 2022-06-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Pyridostatin and Its Derivatives Specifically Binding to G-Quadruplexes.
J.Am.Chem.Soc., 144, 2022
|
|
7X30
 
 | | Capsid structure of Staphylococcus jumbo bacteriophage S6 | | Descriptor: | Hoc-like protein ORF90, Major structural protein ORF12 | | Authors: | Koibuchi, W, Uchiyama, J, Matsuzaki, S, Murata, K, Iwasaki, K, Miyazaki, N. | | Deposit date: | 2022-02-27 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Capsid structure of Staphylococcus jumbo bacteriophage S6
To Be Published
|
|
8S99
 
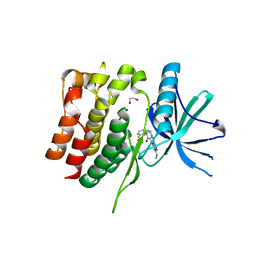 | | Crystal structure of the TYK2 pseudokinase domain in complex with compound 11 | | Descriptor: | (8S)-N-[(1R,2S)-2-fluorocyclopropyl]-5-{[(1M,2'M)-3'-fluoro-2-oxo-2H-[1,2'-bipyridin]-3-yl]amino}-7-(methylamino)pyrazolo[1,5-a]pyrimidine-3-carboxamide, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Toms, A.V, Leit, S, Greenwood, J.R, Carriero, S, Mondal, S, Abel, R, Ashwell, M, Blanchette, H, Boyles, N, Cartwright, M, Collis, A, Feng, S, Ghanakota, P, Harriman, G.C, Hosagrahara, V, Kaila, N, Kapeller, R, Rafi, S, Romero, D.L, Tarantino, P, Timaniya, J, Wester, R.T, Westlin, W, Srivastava, B, Miao, W, Tummino, P, McElwee, J.J, Edmondson, S.D, Massee, C.E. | | Deposit date: | 2023-03-27 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Discovery of a Potent and Selective Tyrosine Kinase 2 Inhibitor: TAK-279.
J.Med.Chem., 66, 2023
|
|
8S9A
 
 | | Crystal structure of the TYK2 pseudokinase domain in complex with TAK-279 | | Descriptor: | (8S)-N-[(1R,2R)-2-methoxycyclobutyl]-7-(methylamino)-5-{[(1P,2'P)-2-oxo-2H-[1,2'-bipyridin]-3-yl]amino}pyrazolo[1,5-a]pyrimidine-3-carboxamide, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Toms, A.V, Leit, S, Greenwood, J.R, Carriero, S, Mondal, S, Abel, R, Ashwell, M, Blanchette, H, Boyles, N, Cartwright, M, Collis, A, Feng, S, Ghanakota, P, Harriman, G.C, Hosagrahara, V, Kaila, N, Kapeller, R, Rafi, S, Romero, D.L, Tarantino, P, Timaniya, J, Wester, R.T, Westlin, W, Srivastava, B, Miao, W, Tummino, P, McElwee, J.J, Edmondson, S.D, Massee, C.E. | | Deposit date: | 2023-03-27 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of a Potent and Selective Tyrosine Kinase 2 Inhibitor: TAK-279.
J.Med.Chem., 66, 2023
|
|
