1O2J
 
 | | Elaborate Manifold of Short Hydrogen Bond Arrays Mediating Binding of Active Site-Directed Serine Protease Inhibitors | | Descriptor: | 2-{5-[AMINO(IMINIO)METHYL]-1H-BENZIMIDAZOL-2-YL}-6-ISOBUTOXYBENZENOLATE, BETA-TRYPSIN, CALCIUM ION, ... | | Authors: | Katz, B.A, Elrod, K, Verner, E, Mackman, R.L, Luong, C, Shrader, W, Sendzik, M, Spencer, J.R, Sprengeler, P.A, Kolesnikov, A, Tai, W.F, Hui, H, Breitenbucher, G, Allen, D, Janc, J. | | Deposit date: | 2003-03-06 | | Release date: | 2003-05-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Elaborate Manifold of Short Hydrogen Bond Arrays Mediating
Binding of Active Site-Directed Serine Protease Inhibitors
J.Mol.Biol., 329, 2003
|
|
1O2H
 
 | | Elaborate Manifold of Short Hydrogen Bond Arrays Mediating Binding of Active Site-Directed Serine Protease Inhibitors | | Descriptor: | 2-{5-[AMINO(IMINIO)METHYL]-1H-INDOL-2-YL}-6-(CYCLOPENTYLOXY)BENZENOLATE, BETA-TRYPSIN, CALCIUM ION | | Authors: | Katz, B.A, Elrod, K, Verner, E, Mackman, R.L, Luong, C, Shrader, W, Sendzik, M, Spencer, J.R, Sprengeler, P.A, Kolesnikov, A, Tai, W.F, Hui, H, Breitenbucher, G, Allen, D, Janc, J. | | Deposit date: | 2003-03-06 | | Release date: | 2003-05-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Elaborate Manifold of Short Hydrogen Bond Arrays Mediating
Binding of Active Site-Directed Serine Protease Inhibitors
J.Mol.Biol., 329, 2003
|
|
1O2M
 
 | | Elaborate Manifold of Short Hydrogen Bond Arrays Mediating Binding of Active Site-Directed Serine Protease Inhibitors | | Descriptor: | 3-{5-[AMINO(IMINIO)METHYL]-6-CHLORO-1H-BENZIMIDAZOL-2-YL}-1,1'-BIPHENYL-2-OLATE, BETA-TRYPSIN, CALCIUM ION, ... | | Authors: | Katz, B.A, Elrod, K, Verner, E, Mackman, R.L, Luong, C, Shrader, W, Sendzik, M, Spencer, J.R, Sprengeler, P.A, Kolesnikov, A, Tai, W.F, Hui, H, Breitenbucher, G, Allen, D, Janc, J. | | Deposit date: | 2003-03-06 | | Release date: | 2003-05-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Elaborate Manifold of Short Hydrogen Bond Arrays Mediating
Binding of Active Site-Directed Serine Protease Inhibitors
J.Mol.Biol., 329, 2003
|
|
1W5C
 
 | | Photosystem II from Thermosynechococcus elongatus | | Descriptor: | 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, BETA-CAROTENE, CHLOROPHYLL A, ... | | Authors: | Biesiadka, J, Loll, B, Kern, J, Irrgang, K.-D, Saenger, W. | | Deposit date: | 2004-08-06 | | Release date: | 2004-12-21 | | Last modified: | 2014-01-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of Cyanobacterial Photosystem II at 3.2 A Resolution: A Closer Look at the Mn- Cluster
Phys.Chem.Chem.Phys., 6, 2004
|
|
1PGV
 
 | | Structural Genomics of Caenorhabditis elegans: tropomodulin C-terminal domain | | Descriptor: | tropomodulin TMD-1 | | Authors: | Symersky, J, Lu, S, Li, S, Chen, L, Meehan, E, Luo, M, Qiu, S, Bunzel, R.J, Luo, D, Arabashi, A, Nagy, L.A, Lin, G, Luan, W.C.-H, Carson, M, Gray, R, Huang, W, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-05-28 | | Release date: | 2003-06-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural genomics of Caenorhabditis elegans: crystal structure of the tropomodulin C-terminal domain
Proteins, 56, 2004
|
|
3ZW1
 
 | | Structure of Bambl lectine in complex with lewix x antigen | | Descriptor: | BAMBL LECTIN, alpha-D-galactopyranose, alpha-L-fucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Audfray, A, Claudinon, J, Abounit, S, Ruvoen-Clouet, N, Larson, G, Wimmerova, M, Lependu, J, Romer, W, Varrot, A, Imberty, A. | | Deposit date: | 2011-07-28 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Deciphering the Glycan Preference of Bacterial Lectins by Glycan Array and Molecular Docking with Validation by Microcalorimetry and Crystallography.
Plos One, 8, 2013
|
|
1O2O
 
 | | Elaborate Manifold of Short Hydrogen Bond Arrays Mediating Binding of Active Site-Directed Serine Protease Inhibitors | | Descriptor: | 2-{5-[AMINO(IMINIO)METHYL]-6-FLUORO-1H-BENZIMIDAZOL-2-YL}-6-ISOBUTOXYBENZENOLATE, BETA-TRYPSIN, CALCIUM ION, ... | | Authors: | Katz, B.A, Elrod, K, Verner, E, Mackman, R.L, Luong, C, Shrader, W, Sendzik, M, Spencer, J.R, Sprengeler, P.A, Kolesnikov, A, Tai, W.F, Hui, H, Breitenbucher, G, Allen, D, Janc, J. | | Deposit date: | 2003-03-06 | | Release date: | 2003-05-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Elaborate Manifold of Short Hydrogen Bond Arrays Mediating
Binding of Active Site-Directed Serine Protease Inhibitors
J.Mol.Biol., 329, 2003
|
|
1NNH
 
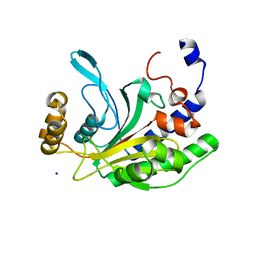 | | Hypothetical protein from Pyrococcus furiosus Pfu-1801964 | | Descriptor: | SODIUM ION, asparaginyl-tRNA synthetase-related peptide | | Authors: | Tempel, W, Liu, Z.-J, Schubot, F.D, Shah, A, Arendall III, W.B, Rose, J.P, Richardson, D.C, Richardson, J.S, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-01-13 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Hypothetical protein from Pyrococcus furiosus Pfu-1801964
To be published
|
|
3URM
 
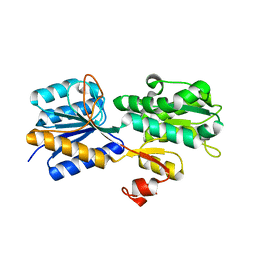 | | Crystal structure of the periplasmic sugar binding protein ChvE | | Descriptor: | Multiple sugar-binding periplasmic receptor ChvE, beta-D-galactopyranose | | Authors: | Hu, X, Zhao, J, Binns, A, Degrado, W. | | Deposit date: | 2011-11-22 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Agrobacterium tumefaciens recognizes its host environment using ChvE to bind diverse plant sugars as virulence signals.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3US8
 
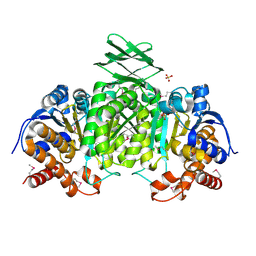 | | Crystal Structure of an isocitrate dehydrogenase from Sinorhizobium meliloti 1021 | | Descriptor: | Isocitrate dehydrogenase [NADP], SULFATE ION | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-11-23 | | Release date: | 2011-12-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of an isocitrate dehydrogenase from Sinorhizobium meliloti 1021
To be Published
|
|
3UUG
 
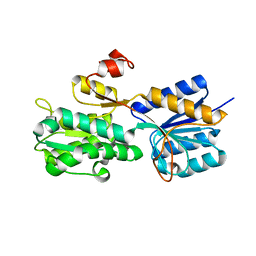 | | Crystal structure of the periplasmic sugar binding protein ChvE | | Descriptor: | Multiple sugar-binding periplasmic receptor ChvE, beta-D-glucopyranuronic acid | | Authors: | Hu, X, Zhao, J, Binns, A, Degrado, W. | | Deposit date: | 2011-11-28 | | Release date: | 2012-11-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Agrobacterium tumefaciens recognizes its host environment using ChvE to bind diverse plant sugars as virulence signals.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3UXY
 
 | | The crystal structure of short chain dehydrogenase from Rhodobacter sphaeroides | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Short-chain dehydrogenase/reductase SDR | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-05 | | Release date: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | The crystal structure of short chain dehydrogenase from Rhodobacter sphaeroides
To be Published
|
|
3V2G
 
 | | Crystal structure of a dehydrogenase/reductase from Sinorhizobium meliloti 1021 | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase, SULFATE ION | | Authors: | Agarwal, R, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-12 | | Release date: | 2012-01-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a dehydrogenase/reductase from Sinorhizobium meliloti 1021
To be Published
|
|
1KU6
 
 | | Fasciculin 2-Mouse Acetylcholinesterase Complex | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Bourne, Y, Burmeister, W, Taylor, P, Marchot, P. | | Deposit date: | 2002-01-21 | | Release date: | 2003-12-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into ligand interactions at the acetylcholinesterase peripheral anionic site.
EMBO J., 22, 2003
|
|
1K8I
 
 | | CRYSTAL STRUCTURE OF MOUSE H2-DM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MHC class II H2-M alpha chain, MHC class II H2-M beta 2 chain | | Authors: | Fremont, D.H, Crawford, F, Marrack, P, Hendrickson, W, Kappler, J. | | Deposit date: | 2001-10-24 | | Release date: | 2001-12-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of mouse H2-M.
Immunity, 9, 1998
|
|
3UN1
 
 | | Crystal structure of an oxidoreductase from Sinorhizobium meliloti 1021 | | Descriptor: | PHOSPHATE ION, Probable oxidoreductase | | Authors: | Agarwal, R, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-11-15 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of an oxidoreductase from Sinorhizobium meliloti 1021
To be Published
|
|
3UTN
 
 | | Crystal structure of Tum1 protein from Saccharomyces cerevisiae | | Descriptor: | DIMETHYL SULFOXIDE, SULFATE ION, Thiosulfate sulfurtransferase TUM1 | | Authors: | Qiu, R, Wang, F, Liu, M, Ji, C, Gong, W. | | Deposit date: | 2011-11-26 | | Release date: | 2012-10-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the Tum1 protein from the yeast Saccharomyces cerevisiae.
Protein Pept.Lett., 19, 2012
|
|
3V2H
 
 | | The crystal structure of D-beta-hydroxybutyrate dehydrogenase from Sinorhizobium meliloti | | Descriptor: | D-beta-hydroxybutyrate dehydrogenase | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-12 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of D-beta-hydroxybutyrate dehydrogenase from Sinorhizobium meliloti
To be Published
|
|
1YDO
 
 | | Crystal Structure of the Bacillis subtilis HMG-CoA Lyase, Northeast Structural Genomics Target SR181. | | Descriptor: | HMG-CoA Lyase, IODIDE ION | | Authors: | Forouhar, F, Hussain, M, Edstrom, W, Vorobiev, S.M, Xiao, R, Ciano, M, Shih, L, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-12-24 | | Release date: | 2005-07-05 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal structures of two bacterial 3-hydroxy-3-methylglutaryl-CoA lyases suggest a common catalytic mechanism among a family of TIM barrel metalloenzymes cleaving carbon-carbon bonds.
J.Biol.Chem., 281, 2006
|
|
3V4C
 
 | | Crystal structure of a semialdehyde dehydrogenase from Sinorhizobium meliloti 1021 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Aldehyde dehydrogenase (NADP+) | | Authors: | Agarwal, R, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-14 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of a semialdehyde dehydrogenase from Sinorhizobium meliloti 1021
To be Published
|
|
1O2I
 
 | | Elaborate Manifold of Short Hydrogen Bond Arrays Mediating Binding of Active Site-Directed Serine Protease Inhibitors | | Descriptor: | 2-{5-[AMINO(IMINIO)METHYL]-1H-BENZIMIDAZOL-2-YL}-6-(CYCLOPENTYLOXY)BENZENOLATE, BETA-TRYPSIN, CALCIUM ION, ... | | Authors: | Katz, B.A, Elrod, K, Verner, E, Mackman, R.L, Luong, C, Shrader, W, Sendzik, M, Spencer, J.R, Sprengeler, P.A, Kolesnikov, A, Tai, W.F, Hui, H, Breitenbucher, G, Allen, D, Janc, J. | | Deposit date: | 2003-03-06 | | Release date: | 2003-05-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Elaborate Manifold of Short Hydrogen Bond Arrays Mediating
Binding of Active Site-Directed Serine Protease Inhibitors
J.Mol.Biol., 329, 2003
|
|
1O2N
 
 | | Elaborate Manifold of Short Hydrogen Bond Arrays Mediating Binding of Active Site-Directed Serine Protease Inhibitors | | Descriptor: | 3-{5-[AMINO(IMINIO)METHYL]-6-CHLORO-1H-BENZIMIDAZOL-2-YL}-1,1'-BIPHENYL-2-OLATE, BETA-TRYPSIN, CALCIUM ION, ... | | Authors: | Katz, B.A, Elrod, K, Verner, E, Mackman, R.L, Luong, C, Shrader, W, Sendzik, M, Spencer, J.R, Sprengeler, P.A, Kolesnikov, A, Tai, W.F, Hui, H, Breitenbucher, G, Allen, D, Janc, J. | | Deposit date: | 2003-03-06 | | Release date: | 2003-05-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Elaborate Manifold of Short Hydrogen Bond Arrays Mediating
Binding of Active Site-Directed Serine Protease Inhibitors
J.Mol.Biol., 329, 2003
|
|
1O2L
 
 | | Elaborate Manifold of Short Hydrogen Bond Arrays Mediating Binding of Active Site-Directed Serine Protease Inhibitors | | Descriptor: | 3-{5-[AMINO(IMINIO)METHYL]-6-CHLORO-1H-BENZIMIDAZOL-2-YL}-1,1'-BIPHENYL-2-OLATE, BETA-TRYPSIN, CALCIUM ION | | Authors: | Katz, B.A, Elrod, K, Verner, E, Mackman, R.L, Luong, C, Shrader, W, Sendzik, M, Spencer, J.R, Sprengeler, P.A, Kolesnikov, A, Tai, W.F, Hui, H, Breitenbucher, G, Allen, D, Janc, J. | | Deposit date: | 2003-03-06 | | Release date: | 2003-05-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Elaborate Manifold of Short Hydrogen Bond Arrays Mediating
Binding of Active Site-Directed Serine Protease Inhibitors
J.Mol.Biol., 329, 2003
|
|
1LPF
 
 | |
4ABN
 
 | | Crystal structure of full length mouse Strap (TTC5) | | Descriptor: | 1,2-ETHANEDIOL, TETRATRICOPEPTIDE REPEAT PROTEIN 5 | | Authors: | Pike, A.C.W, Bullock, A.N, Kleinekofort, W, Zimmermann, T, Burgess-Brown, N, Sharpe, T.D, Thangaratnarajah, C, Keates, T, Ugochukwu, E, Bunkoczi, G, Uppenberg, J, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, La Thangue, N.B, Knapp, S. | | Deposit date: | 2011-12-09 | | Release date: | 2012-01-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The P53 Cofactor Strap Exhibits an Unexpected Tpr Motif and Oligonucleotide-Binding (Ob)-Fold Structure.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
