8J0D
 
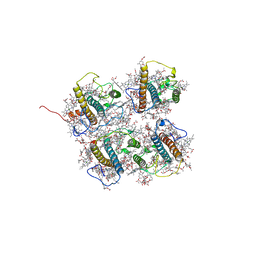 | | FCP heterodimer, Lhca2, and Lhcf5 together as the M1 side binds to the PSII core in the diatom Thalassiosira pseudonana | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Li, Z, Feng, Y, Wang, W, Shen, J.R. | | Deposit date: | 2023-04-10 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structure of a diatom photosystem II supercomplex containing a member of Lhcx family and dimeric FCPII.
Sci Adv, 9, 2023
|
|
3TAR
 
 | |
8JHF
 
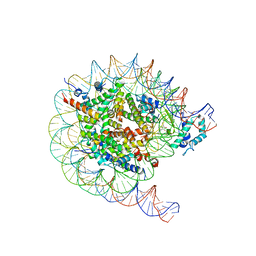 | | Native SUV420H1 bound to 167-bp nucleosome | | Descriptor: | DNA (160-MER), Histone H2A.Z, Histone H2B type 1-K, ... | | Authors: | Lin, F, Li, W. | | Deposit date: | 2023-05-23 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structural basis of nucleosomal H4K20 recognition and methylation by SUV420H1 methyltransferase.
Cell Discov, 9, 2023
|
|
3SZL
 
 | | IspH:Ligand Mutants - wt 70sec | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IRON/SULFUR CLUSTER | | Authors: | Span, I, Graewert, T, Bacher, A, Eisenreich, W, Groll, M. | | Deposit date: | 2011-07-19 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of Mutant IspH Proteins Reveal a Rotation of the Substrate's Hydroxymethyl Group during Catalysis.
J.Mol.Biol., 416, 2012
|
|
8JHG
 
 | | Native SUV420H1 bound to 167-bp nucleosome | | Descriptor: | DNA (160-MER), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | Authors: | Lin, F, Li, W. | | Deposit date: | 2023-05-23 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural basis of nucleosomal H4K20 recognition and methylation by SUV420H1 methyltransferase.
Cell Discov, 9, 2023
|
|
4UUG
 
 | | The (R)-selective amine transaminase from Aspergillus fumigatus with inhibitor bound | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 3-[O-PHOSPHONOPYRIDOXYL]--AMINO-BENZOIC ACID, ... | | Authors: | Thomsen, M, Hinrichs, W. | | Deposit date: | 2014-07-28 | | Release date: | 2014-11-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Biochemical Characterization of the Dual Substrate Recognition of the (R)-Selective Amine Transaminase from Aspergillus Fumigatus
FEBS J., 282, 2015
|
|
3T4D
 
 | | Crystal Structure of NaK2K Channel Y55F Mutant | | Descriptor: | POTASSIUM ION, Potassium channel protein | | Authors: | Sauer, D.B, Zeng, W, Raghunathan, S, Jiang, Y. | | Deposit date: | 2011-07-25 | | Release date: | 2011-10-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Protein interactions central to stabilizing the K+ channel selectivity filter in a four-sited configuration for selective K+ permeation.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3SZO
 
 | | IspH:HMBPP complex after 3 minutes X-ray pre-exposure | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IRON/SULFUR CLUSTER | | Authors: | Span, I, Graewert, T, Bacher, A, Eisenreich, W, Groll, M. | | Deposit date: | 2011-07-19 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of Mutant IspH Proteins Reveal a Rotation of the Substrate's Hydroxymethyl Group during Catalysis.
J.Mol.Biol., 416, 2012
|
|
3ST0
 
 | | Engineered medium-affinity halide-binding protein derived from YFP: halide-free | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Green fluorescent protein | | Authors: | Wang, W, Grimley, J.S, Beese, L.S, Hellinga, H.W. | | Deposit date: | 2011-07-08 | | Release date: | 2012-07-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Visualization of Synaptic Inhibition with an Optogenetic Sensor Developed by Cell-Free Protein Engineering Automation.
J.Neurosci., 33, 2013
|
|
3TC5
 
 | | Selective targeting of disease-relevant protein binding domains by O-phosphorylated natural product derivatives | | Descriptor: | (11alpha,16alpha)-9-fluoro-11,17-dihydroxy-16-methyl-3,20-dioxopregna-1,4-dien-21-yl dihydrogen phosphate, HEXAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Graeber, M, Janczyk, W, Sperl, B, Elumalai, N, Kozany, C, Hausch, F, Holak, T.A, Berg, T. | | Deposit date: | 2011-08-08 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Selective targeting of disease-relevant protein binding domains by o-phosphorylated natural product derivatives.
Acs Chem.Biol., 6, 2011
|
|
9BRR
 
 | | Intact V-ATPase State 3 in synaptophysin knock-out isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, V-type proton ATPase 116 kDa subunit a 1, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
9RNT
 
 | |
9BRA
 
 | | Intact V-ATPase State 2 and synaptophysin complex in mouse brain isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, Synaptophysin, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
9BRT
 
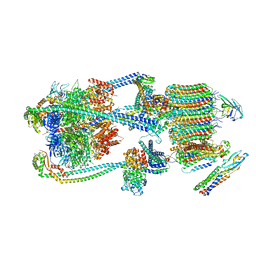 | | Intact V-ATPase State 1 and synaptophysin complex in mouse brain isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, Synaptophysin, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
9BRZ
 
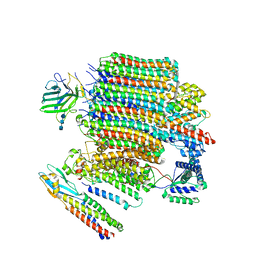 | | V0-only V-ATPase and synaptophysin complex in mouse brain isolated synaptic vesicles | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-12 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
4MDC
 
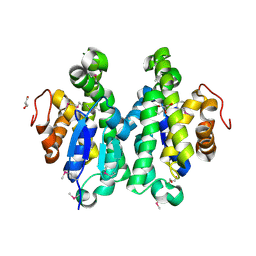 | | Crystal structure of glutathione S-transferase from Sinorhizobium meliloti 1021, NYSGRC target 021389 | | Descriptor: | GLYCEROL, Putative glutathione S-transferase | | Authors: | Shabalin, I.G, Bacal, P, Cooper, D.R, Stead, M, Ahmed, M, Hammonds, J, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-22 | | Release date: | 2013-09-04 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of glutathione S-transferase from Sinorhizobium meliloti 1021
To be Published
|
|
9BRS
 
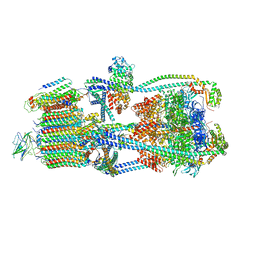 | | Intact V-ATPase State 2 in synaptophysin knock-out isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, V-type proton ATPase 116 kDa subunit a 1, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
9BRU
 
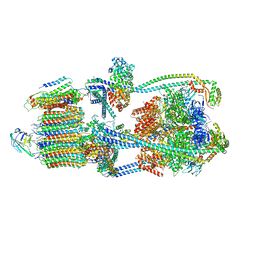 | | Intact V-ATPase State 1 in synaptophysin knock-out isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, V-type proton ATPase 116 kDa subunit a 1, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
4M7Y
 
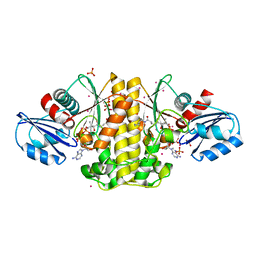 | | Staphylococcus aureus Type II pantothenate kinase in complex with a pantothenate analog | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-N-pentyl-beta-alaninamide, PHOSPHATE ION, ... | | Authors: | Mottaghi, K, Hong, B, Tempel, W, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-08-12 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Potent Pantothenamide Inhibitors of Staphylococcus aureus Pantothenate Kinase through a Minimal SAR Study: Inhibition Is Due to Trapping of the Product.
ACS Infect Dis, 2, 2016
|
|
4LVD
 
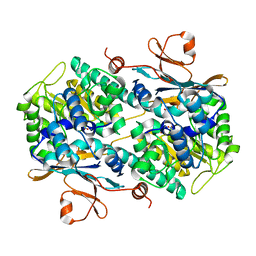 | | Fragment-based Identification of Amides Derived From trans-2-(Pyridin-3-yl)cyclopropanecarboxylic Acid as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | 1,2-ETHANEDIOL, N-(4-nitrophenyl)cyclopropanecarboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Giannetti, A.M, Zheng, X, Skelton, N, Wang, W, Bravo, B, Feng, Y, Gunzner-Toste, J, Ho, Y, Hua, R, Wang, C, Zhao, Q, Liederer, B.M, Liu, Y, O'Brien, T, Oeh, J, Sampath, D, Shen, Y, Wang, L, Wu, H, Xiao, Y, Yuen, P, Zak, M, Zhao, G, Dragovich, P.S. | | Deposit date: | 2013-07-26 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification of amides derived from 1H-pyrazolo[3,4-b]pyridine-5-carboxylic acid as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4LX2
 
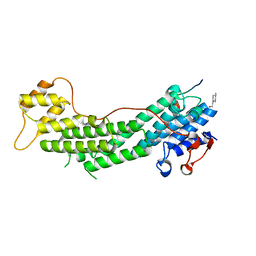 | | Crystal structure of Myo5a globular tail domain in complex with melanophilin GTBD | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Melanophilin, Unconventional myosin-Va | | Authors: | Pylypenko, O, Attanda, W, Gauquelin, C, Houdusse, A. | | Deposit date: | 2013-07-29 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of myosin V Rab GTPase-dependent cargo recognition.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
9BRQ
 
 | | Intact V-ATPase State 3 and synaptophysin complex in mouse brain isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, Synaptophysin, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
9BRY
 
 | | V0-only V-ATPase in synaptophysin gene knock-out mouse brain isolated synaptic vesicles | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-12 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
4M0Q
 
 | | Ebola virus VP24 structure | | Descriptor: | Membrane-associated protein VP24 | | Authors: | Xu, W, Leung, D.W, Borek, D, Amarasinghe, G.K. | | Deposit date: | 2013-08-01 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.921 Å) | | Cite: | The Marburg Virus VP24 Protein Interacts with Keap1 to Activate the Cytoprotective Antioxidant Response Pathway.
Cell Rep, 6, 2014
|
|
4MAZ
 
 | | The Structure of MalL mutant enzyme V200S from Bacillus subtilus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Hobbs, J.K, Jiao, W, Easter, A.D, Parker, E.J, Schipper, L.A, Arcus, V.L. | | Deposit date: | 2013-08-18 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Change in heat capacity for enzyme catalysis determines temperature dependence of enzyme catalyzed rates.
Acs Chem.Biol., 8, 2013
|
|
