3RNO
 
 | | Crystal Structure of Mouse Apolipoprotein A-I Binding Protein in Complex with NADP. | | Descriptor: | Apolipoprotein A-I-binding protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Shumilin, I.A, Jha, K.N, Cymborowski, M, Herr, J.C, Minor, W. | | Deposit date: | 2011-04-22 | | Release date: | 2012-05-09 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RTE
 
 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima soaked with NADP and ATP. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-05-03 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3ROZ
 
 | | Crystal Structure of Mouse Apolipoprotein A-I Binding Protein in Complex with Nicotinamide | | Descriptor: | Apolipoprotein A-I-binding protein, NICOTINAMIDE, SULFATE ION | | Authors: | Shumilin, I.A, Jha, K.N, Cymborowski, M, Herr, J.C, Minor, W. | | Deposit date: | 2011-04-26 | | Release date: | 2012-07-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RQ8
 
 | | Crystal Structure of ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Bacillus subtilis soaked with P1,P5-Di(adenosine-5') pentaphosphate | | Descriptor: | ADP/ATP-dependent NAD(P)H-hydrate dehydratase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION | | Authors: | Shumilin, I.A, Cymborowski, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-27 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RS9
 
 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima in complex with P1,P3-Di(adenosine-5') triphosphate | | Descriptor: | BIS(ADENOSINE)-5'-TRIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-05-02 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RSQ
 
 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima soaked with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, BETA-6-HYDROXY-1,4,5,6-TETRHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, POTASSIUM ION, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-05-02 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.054 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RTG
 
 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima soaked with Coenzyme A and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, COENZYME A, MAGNESIUM ION, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-05-03 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RU7
 
 | |
3RU9
 
 | |
4H34
 
 | | Crystal structure of the tyrosine phosphatase SHP-2 with Q506P mutation | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Qiu, W, Lin, A, Hutchinson, A, Romanov, V, Ruzanov, M, Thompson, C, Lam, K, Kisselman, G, Battaile, K, Chirgadze, N.Y. | | Deposit date: | 2012-09-13 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the tyrosine phosphatase SHP-2 with Q506P mutation
To be Published
|
|
3RUH
 
 | |
3RUD
 
 | |
7LYZ
 
 | | PROTEIN MODEL BUILDING BY THE USE OF A CONSTRAINED-RESTRAINED LEAST-SQUARES PROCEDURE | | Descriptor: | HEN EGG WHITE LYSOZYME | | Authors: | Moult, J, Yonath, A, Sussman, J, Herzberg, O, Podjarny, A, Traub, W. | | Deposit date: | 1977-05-06 | | Release date: | 1977-06-20 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Protein Model Building by the Use of a Constrained-Restrained Least-Squares Procedure
J.Appl.Crystallogr., 16, 1983
|
|
3RXW
 
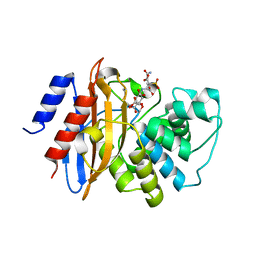 | | KPC-2 carbapenemase in complex with PSR3-226 | | Descriptor: | (2S,3R)-4-(2-amino-2-oxoethoxy)-3-(dihydroxy-lambda~4~-sulfanyl)-3-methyl-4-oxo-2-{[(1E)-3-oxoprop-1-en-1-yl]amino}butanoic acid, CITRIC ACID, Carbepenem-hydrolyzing beta-lactamase KPC | | Authors: | Ke, W, van den Akker, F. | | Deposit date: | 2011-05-10 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystal structures of KPC-2 {beta}-lactamase in complex with 3-nitrophenyl boronic acid and the penam sulfone PSR-3-226.
Antimicrob.Agents Chemother., 56, 2012
|
|
4JKV
 
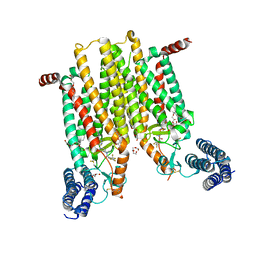 | | Structure of the human smoothened 7TM receptor in complex with an antitumor agent | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-fluoro-N-methyl-N-{1-[4-(1-methyl-1H-pyrazol-5-yl)phthalazin-1-yl]piperidin-4-yl}-2-(trifluoromethyl)benzamide, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wang, C, Wu, H, Katritch, V, Han, G.W, Huang, X, Liu, W, Siu, F.Y, Roth, B.L, Cherezov, V, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2013-03-11 | | Release date: | 2013-04-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of the human smoothened receptor bound to an antitumour agent.
Nature, 497, 2013
|
|
3RU3
 
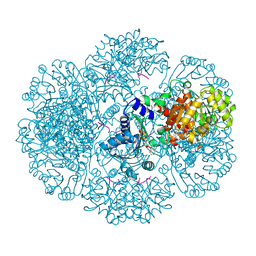 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima soaked with NADPH and ATP. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, BETA-6-HYDROXY-1,4,5,6-TETRAHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE PHOSPHATE, MAGNESIUM ION, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-05-04 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RUC
 
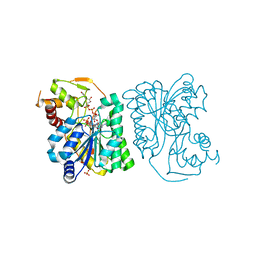 | |
4JNM
 
 | | Discovery of Potent and Efficacious Urea-containing Nicotinamide Phosphoribosyltransferase (NAMPT) Inhibitors with Reduced CYP2C9 Inhibition Properties | | Descriptor: | 1,2-ETHANEDIOL, 1-[(6-aminopyridin-3-yl)methyl]-3-[4-(phenylsulfonyl)phenyl]urea, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Gunzner-Toste, J, Zhao, G, Bauer, P, Baumeister, T, Buckmelter, A.J, Caligiuri, M, Clodfelter, K.H, Fu, B, Han, B, Ho, Y, Kley, N, Liederer, B, Lin, J, Mukadam, S, O'Brien, T, Reynolds, D.J, Sharma, G, Skelton, N, Smith, C.C, Oh, A, Wang, W, Wang, Z, Xiao, Y, Yuen, P, Zak, M, Zhang, L, Zheng, X, Bair, K.W, Dragovich, P.S. | | Deposit date: | 2013-03-15 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of potent and efficacious urea-containing nicotinamide phosphoribosyltransferase (NAMPT) inhibitors with reduced CYP2C9 inhibition properties.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3RZH
 
 | | Duplex Interrogation by a Direct DNA Repair Protein in the Search of Damage | | Descriptor: | 5'-D(*CP*TP*GP*TP*CP*TP*(ME6)P*AP*CP*TP*GP*CP*G)-3', 5'-D(*TP*CP*GP*CP*AP*GP*TP*GP*AP*GP*AP*CP*A)-3', Alpha-ketoglutarate-dependent dioxygenase alkB homolog 2, ... | | Authors: | Yi, C, Chen, B, Qi, B, Zhang, W, Jia, G, Zhang, L, Li, C, Dinner, A, Yang, C, He, C. | | Deposit date: | 2011-05-11 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Duplex interrogation by a direct DNA repair protein in search of base damage
Nat.Struct.Mol.Biol., 19, 2012
|
|
3RY6
 
 | | Complex of fcgammariia (CD32) and the FC of human IGG1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ig gamma-1 chain C region, ... | | Authors: | Ramsland, P.A, Farrugia, W, Scott, A.M, Hogarth, P.M. | | Deposit date: | 2011-05-11 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural Basis for Fc{gamma}RIIa Recognition of Human IgG and Formation of Inflammatory Signaling Complexes.
J.Immunol., 187, 2011
|
|
3RVT
 
 | | Structure of 4C1 Fab in P212121 space group | | Descriptor: | Fab fragment of 4C1 antibody - heavy chain, Fab fragment of 4C1 antibody - light chain | | Authors: | Chruszcz, M, Vailes, L.D, Chapman, M.D, Pomes, A, Minor, W. | | Deposit date: | 2011-05-06 | | Release date: | 2012-01-11 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Molecular determinants for antibody binding on group 1 house dust mite allergens.
J.Biol.Chem., 287, 2012
|
|
4JT3
 
 | | Crystal Structure of TTK kinase domain with an inhibitor: 400740 | | Descriptor: | 1,2-ETHANEDIOL, 2-phenyl-N-[3-(3-sulfamoylphenyl)-2H-indazol-5-yl]acetamide, Dual specificity protein kinase TTK, ... | | Authors: | Qiu, W, Harris-Brandts, M, Feher, M, Awrey, D.E, Chirgadze, N.Y. | | Deposit date: | 2013-03-22 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of TTK kinase domain with an inhibitor: 400740
To be Published
|
|
4JSB
 
 | | Crystal structure of Tfu_1878, a putative enoyl-CoA hydratase from Thermobifida fusca YX | | Descriptor: | Enoyl-CoA hydratase, SULFATE ION | | Authors: | Mikolajczak, K, Porebski, P.J, Cooper, D.R, Ahmed, M, Stead, M, Hillerich, B, Seidel, R, Zimmerman, M, Bonanno, J.B, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-03-22 | | Release date: | 2013-06-19 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure of Tfu_1878, a putative enoyl-CoA hydratase from Thermobifida fusca YX
TO BE PUBLISHED
|
|
3RY4
 
 | |
4JTV
 
 | | Crystal structure of 2009 pandemic influenza virus hemagglutinin complexed with human receptor analogue LSTc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Zhang, W, Shi, Y, Qi, J, Gao, F, Li, Q, Fan, Z, Yan, J, Gao, G.F. | | Deposit date: | 2013-03-24 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.997 Å) | | Cite: | Molecular basis of the receptor binding specificity switch of the hemagglutinins from both the 1918 and 2009 pandemic influenza A viruses by a D225G substitution
J.Virol., 87, 2013
|
|
