6PMJ
 
 | | Sigm28-transcription initiation complex with specific promoter at the state 2 | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Liu, B, Shi, W. | | Deposit date: | 2019-07-02 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Structural basis of bacterial sigma28-mediated transcription reveals roles of the RNA polymerase zinc-binding domain.
Embo J., 39, 2020
|
|
5JBX
 
 | | Crystal structure of LiuC in complex with coenzyme A and malonic acid | | Descriptor: | 3-hydroxybutyryl-CoA dehydratase, COENZYME A, MALONATE ION | | Authors: | Bock, T, Reichelt, J, Mueller, R, Blankenfeldt, W. | | Deposit date: | 2016-04-14 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The Structure of LiuC, a 3-Hydroxy-3-Methylglutaconyl CoA Dehydratase Involved in Isovaleryl-CoA Biosynthesis in Myxococcus xanthus, Reveals Insights into Specificity and Catalysis.
Chembiochem, 17, 2016
|
|
6F5M
 
 | | Crystal structure of highly glycosylated human leukocyte elastase in complex with a thiazolidinedione inhibitor | | Descriptor: | 5-[[4-[[(2~{S})-4-methyl-1-oxidanylidene-1-[(2-propylphenyl)amino]pentan-2-yl]carbamoyl]phenyl]methyl]-2-oxidanylidene-1,3-thiazol-1-ium-4-olate, ACETATE ION, Neutrophil elastase, ... | | Authors: | Hochscherf, J, Pietsch, M, Tieu, W, Kuan, K, Hautmann, S, Abell, A, Guetschow, M, Niefind, K. | | Deposit date: | 2017-12-01 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of highly glycosylated human leukocyte elastase in complex with an S2' site binding inhibitor.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
5JES
 
 | | Human carbonic anhydrase II (V121T) complexed with benzo[d]thiazole-2-sulfonamide | | Descriptor: | 1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Fox, J.M, Kang, K, Sastry, M, Sherman, W, Sankaran, B, Zwart, P.H, Whitesides, G.M. | | Deposit date: | 2016-04-18 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.205 Å) | | Cite: | Water-Restructuring Mutations Can Reverse the Thermodynamic Signature of Ligand Binding to Human Carbonic Anhydrase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5JGK
 
 | | Crystal structure of GtmA in complex with SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, UbiE/COQ5 family methyltransferase, ... | | Authors: | Dolan, S.K, Bock, T, Hering, V, Jones, G.W, Blankenfeldt, W, Dolye, S. | | Deposit date: | 2016-04-20 | | Release date: | 2017-03-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structural, mechanistic and functional insight into gliotoxinbis-thiomethylation inAspergillus fumigatus.
Open Biol, 7, 2017
|
|
5IWE
 
 | | E45Q mutant of phenazine biosynthesis protein PhzF in complex with (5R,6R)-6-azaniumyl-5-ethoxycyclohexa-1,3-diene-1-carboxylate | | Descriptor: | (5R,6R)-6-azaniumyl-5-ethoxycyclohexa-1,3-diene-1-carboxylate, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Diederich, C, Blankenfeldt, W. | | Deposit date: | 2016-03-22 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Mechanisms and Specificity of Phenazine Biosynthesis Protein PhzF.
Sci Rep, 7, 2017
|
|
1QH4
 
 | | CRYSTAL STRUCTURE OF CHICKEN BRAIN-TYPE CREATINE KINASE AT 1.41 ANGSTROM RESOLUTION | | Descriptor: | ACETATE ION, CALCIUM ION, CREATINE KINASE | | Authors: | Eder, M, Schlattner, U, Becker, A, Wallimann, T, Kabsch, W, Fritz-Wolf, K. | | Deposit date: | 1999-05-11 | | Release date: | 1999-11-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystal structure of brain-type creatine kinase at 1.41 A resolution.
Protein Sci., 8, 1999
|
|
5J1W
 
 | | Crystal structure of human CLK1 in complex with pyrido[3,4-g]quinazoline derivative ZW31 (compound 14) | | Descriptor: | Dual specificity protein kinase CLK1, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Chaikuad, A, Esvan, Y.J, Zeinyeh, W, Boibessot, T, Nauton, L, Thery, V, Loaec, N, Meijer, L, Giraud, F, Moreau, P, Anizon, F, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-03-29 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Discovery of pyrido[3,4-g]quinazoline derivatives as CMGC family protein kinase inhibitors: Design, synthesis, inhibitory potency and X-ray co-crystal structure.
Eur.J.Med.Chem., 118, 2016
|
|
5J28
 
 | | Ki67-PP1g (protein phosphatase 1, gamma isoform) holoenzyme complex | | Descriptor: | Antigen KI-67, MALONATE ION, SODIUM ION, ... | | Authors: | Kumar, G.S, Peti, W, Page, R. | | Deposit date: | 2016-03-29 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Ki-67 and RepoMan mitotic phosphatases assemble via an identical, yet novel mechanism.
Elife, 5, 2016
|
|
6FKH
 
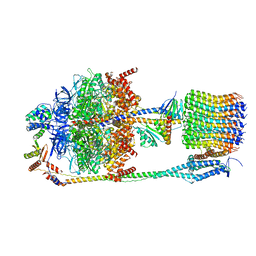 | | Chloroplast F1Fo conformation 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase delta chain, ... | | Authors: | Hahn, A, Vonck, J, Mills, D.J, Meier, T, Kuehlbrandt, W. | | Deposit date: | 2018-01-24 | | Release date: | 2018-05-23 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure, mechanism, and regulation of the chloroplast ATP synthase.
Science, 360, 2018
|
|
5J41
 
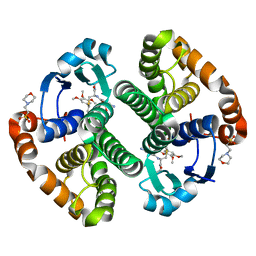 | | Glutathione S-transferase bound with hydrolyzed Piperlongumine | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-(3,4,5-trimethoxyphenyl)propanoic acid, GLUTATHIONE, ... | | Authors: | Harshbarger, W, Gondi, S, Ficarro, S, Hunter, J, Udayakumar, D, Gurbani, D, Marto, J, Westover, K. | | Deposit date: | 2016-03-31 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.19035351 Å) | | Cite: | Structural and Biochemical Analyses Reveal the Mechanism of Glutathione S-Transferase Pi 1 Inhibition by the Anti-cancer Compound Piperlongumine.
J. Biol. Chem., 292, 2017
|
|
5J23
 
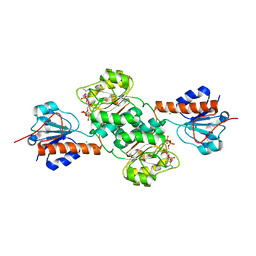 | | Crystal structure of NADPH-dependent glyoxylate/hydroxypyruvate reductase SMc04462 (SmGhrB) from Sinorhizobium meliloti in complex with 2'-phospho-ADP-ribose | | Descriptor: | 2-hydroxyacid dehydrogenase, ACETATE ION, CHLORIDE ION, ... | | Authors: | Shabalin, I.G, Gasiorowska, O.A, Handing, K.B, Bonanno, J, Kutner, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2016-03-29 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural, Biochemical, and Evolutionary Characterizations of Glyoxylate/Hydroxypyruvate Reductases Show Their Division into Two Distinct Subfamilies.
Biochemistry, 57, 2018
|
|
5JEG
 
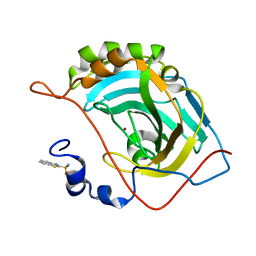 | | Human carbonic anhydrase II (V121I) complexed with benzo[d]thiazole-2-sulfonamide | | Descriptor: | 1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Fox, J.M, Kang, K, Sastry, M, Sherman, W, Sankaran, B, Zwart, P.H, Whitesides, G.M. | | Deposit date: | 2016-04-18 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Water-Restructuring Mutations Can Reverse the Thermodynamic Signature of Ligand Binding to Human Carbonic Anhydrase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5JEP
 
 | | Human carbonic anhydrase II (T199S) complexed with benzo[d]thiazole-2-sulfonamide | | Descriptor: | 1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Fox, J.M, Kang, K, Sastry, M, Sherman, W, Sankaran, B, Zwart, P.H, Whitesides, G.M. | | Deposit date: | 2016-04-18 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Water-Restructuring Mutations Can Reverse the Thermodynamic Signature of Ligand Binding to Human Carbonic Anhydrase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5J3U
 
 | | Co-crystal structure of the regulatory domain of Toxoplasma gondii PKA with cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, GLYCEROL, Protein Kinase A | | Authors: | El Bakkouri, M, Walker, J.R, Tempel, W, Loppnau, P, Graslund, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Lin, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-03-31 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Co-crystal structure of the regulatory domain of Toxoplasma gondii PKA with cAMP
To Be Published
|
|
5JH9
 
 | | Crystal structure of prApe1 | | Descriptor: | CACODYLATE ION, Vacuolar aminopeptidase 1, ZINC ION | | Authors: | Noda, N.N, Adachi, W, Inagaki, F. | | Deposit date: | 2016-04-20 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Receptor-Mediated Selective Autophagy of Aminopeptidase I Aggregates
Cell Rep, 16, 2016
|
|
6FKI
 
 | | Chloroplast F1Fo conformation 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase delta chain, ... | | Authors: | Hahn, A, Vonck, J, Mills, D.J, Meier, T, Kuehlbrandt, W. | | Deposit date: | 2018-01-24 | | Release date: | 2018-05-23 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure, mechanism, and regulation of the chloroplast ATP synthase.
Science, 360, 2018
|
|
5JG3
 
 | | Human carbonic anhydrase II (121T/N67Q) complexed with benzo[d]thiazole-2-sulfonamide | | Descriptor: | 1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Fox, J.M, Kang, K, Sastry, M, Sherman, W, Sankaran, B, Zwart, P.H, Whitesides, G.M. | | Deposit date: | 2016-04-19 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Water-Restructuring Mutations Can Reverse the Thermodynamic Signature of Ligand Binding to Human Carbonic Anhydrase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5JGS
 
 | | Human carbonic anhydrase II (F131Y/L198A) complexed with benzo[d]thiazole-2-sulfonamide | | Descriptor: | 1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Fox, J.M, Kang, K, Sastry, M, Sherman, W, Sankaran, B, Zwart, P.H, Whitesides, G.M. | | Deposit date: | 2016-04-20 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Water-Restructuring Mutations Can Reverse the Thermodynamic Signature of Ligand Binding to Human Carbonic Anhydrase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5JJX
 
 | | Crystal structure of the HAT domain of sart3 | | Descriptor: | CHLORIDE ION, Squamous cell carcinoma antigen recognized by T-cells 3, UNKNOWN ATOM OR ION | | Authors: | DONG, A, ZHANG, Q, TEMPEL, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, TONG, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-04-25 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the HAT domain of sart3
to be published
|
|
6FKF
 
 | | Chloroplast F1Fo conformation 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase delta chain, ... | | Authors: | Hahn, A, Vonck, J, Mills, D.J, Meier, T, Kuehlbrandt, W. | | Deposit date: | 2018-01-24 | | Release date: | 2018-05-23 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure, mechanism, and regulation of the chloroplast ATP synthase.
Science, 360, 2018
|
|
3BP7
 
 | | The high resolution crystal structure of HLA-B*2709 in complex with a Cathepsin A signal sequence peptide, pCatA | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Kumar, P, Vahedi-Faridi, A, Saenger, W, Uchanska-Ziegler, B, Ziegler, A. | | Deposit date: | 2007-12-18 | | Release date: | 2008-12-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for T cell alloreactivity among three HLA-B14 and HLA-B27 antigens
J.Biol.Chem., 284, 2009
|
|
9FYP
 
 | | Cryo EM structure of the type 3B polymorph of alpha-synuclein at low pH. | | Descriptor: | Alpha-synuclein, CHLORIDE ION | | Authors: | Frey, L, Qureshi, B.M, Kwiatkowski, W, Rhyner, D, Greenwald, J, Riek, R. | | Deposit date: | 2024-07-03 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.23 Å) | | Cite: | On the pH-dependence of alpha-synuclein amyloid polymorphism and the role of secondary nucleation in seed-based amyloid propagation
Elife, 2024
|
|
9FT1
 
 | | Yeast 20S proteasome in complex with epoxyketone inhibitor 13 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Maurits, E, Huber, E.M, Dekker, P.M, Wang, X, Heinemeyer, W, Florea, B.I, Groll, M, Overkleeft, H.S. | | Deposit date: | 2024-06-23 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based design of peptide epoxyketones selectively targeting the three human immunoproteasome active sites
to be published
|
|
8YM1
 
 | | Structure of SADS-CoV Virus Nucleocapsid Protein | | Descriptor: | nucleocapsid phosphoprotein | | Authors: | Zhang, Y, Wu, F, Xu, W. | | Deposit date: | 2024-03-08 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unraveling the assembly mechanism of SADS-CoV virus nucleocapsid protein: insights from RNA binding, dimerization, and epitope diversity profiling
J.Virol., 2024
|
|
