4DFM
 
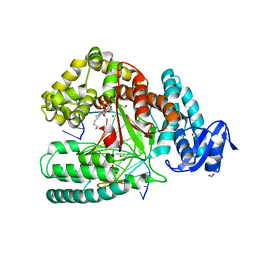 | | Crystal structure of the large fragment of DNA polymerase I from Thermus aquaticus in ternary complex with 5-(aminopentinyl)-2-dCTP | | Descriptor: | 1,2-ETHANEDIOL, 5'-d(AAAGCGCGCCGTGGTC)-3', 5'-d(GACCACGGCGC ddG)-3', ... | | Authors: | Bergen, K, Steck, A, Struett, S, Baccaro, A, Welte, W, Diederichs, K, Marx, A. | | Deposit date: | 2012-01-24 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.886 Å) | | Cite: | Structures of KlenTaq DNA Polymerase Caught While Incorporating C5-Modified Pyrimidine and C7-Modified 7-Deazapurine Nucleoside Triphosphates.
J.Am.Chem.Soc., 134, 2012
|
|
4I6Z
 
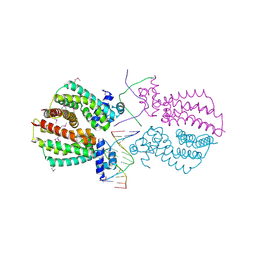 | | Crystal structure of the transcriptional regulator TM1030 with 24bp DNA oligonucleotide | | Descriptor: | DNA OLIGONUCLEOTIDE, Transcriptional regulator, TetR family | | Authors: | Koclega, K.D, Chruszcz, M, Cooper, D.R, Petkowski, J.J, Tkaczuk, K.L, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-11-30 | | Release date: | 2013-01-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the transcriptional regulator TM1030 with 24bp DNA oligonucleotide
To be Published
|
|
4DZD
 
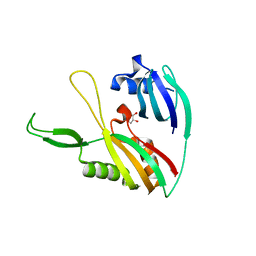 | |
4IAR
 
 | | Crystal structure of the chimeric protein of 5-HT1B-BRIL in complex with ergotamine (PSI Community Target) | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Chimera protein of human 5-hydroxytryptamine receptor 1B and E. Coli soluble cytochrome b562, Ergotamine | | Authors: | Wang, C, Jiang, Y, Ma, J, Wu, H, Wacker, D, Katritch, V, Han, G.W, Liu, W, Huang, X, Vardy, E, McCorvy, J.D, Gao, X, Zhou, E.X, Melcher, K, Zhang, C, Bai, F, Yang, H, Yang, L, Jiang, H, Roth, B.L, Cherezov, V, Stevens, R.C, Xu, H.E, GPCR Network (GPCR) | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for molecular recognition at serotonin receptors.
Science, 340, 2013
|
|
8UEU
 
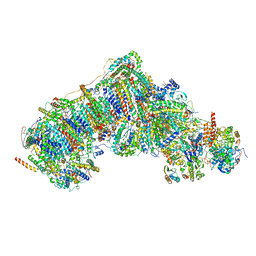 | | In-situ complex I, Deactive class03 | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ... | | Authors: | Zheng, W, Zhu, J, Zhang, K. | | Deposit date: | 2023-10-02 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | High-resolution in situ structures of mammalian respiratory supercomplexes.
Nature, 2024
|
|
8UGE
 
 | | In-situ complex III, state II | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CARDIOLIPIN, ... | | Authors: | Zheng, W, Zhang, K, Zhu, J. | | Deposit date: | 2023-10-05 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | High-resolution in situ structures of mammalian respiratory supercomplexes.
Nature, 2024
|
|
4HXB
 
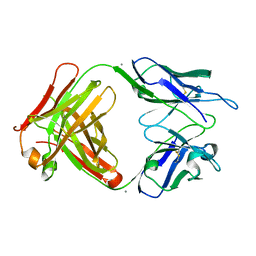 | | Crystal structure of 6B9 FAB | | Descriptor: | 6B9 FAB HEAVY CHAIN, 6B9 FAB LIGHT CHAIN, CALCIUM ION | | Authors: | Johal, A.R, Jarrell, H.C, Khieu, N.H, Letts, J.A, Landry, R.C, Jachymek, W, Yang, Q, Jennings, H.J, Brisson, J.R, Evans, S.V. | | Deposit date: | 2012-11-09 | | Release date: | 2013-07-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The antigen-binding site of an N-propionylated polysialic acid-specific antibody protective against group B meningococci is consistent with extended epitopes.
Glycobiology, 23, 2013
|
|
4D5C
 
 | |
4D0A
 
 | |
4D3Y
 
 | |
4D0Q
 
 | | Hyaluronan Binding Module of the Streptococcal Pneumoniae Hyaluronate Lyase | | Descriptor: | 1,2-ETHANEDIOL, HYALURONATE LYASE | | Authors: | Suits, M.D.L, Pluvinage, B, Law, A, Liu, Y, Palma, A.S, Chai, W, Feizi, T, Boraston, A.B. | | Deposit date: | 2014-04-29 | | Release date: | 2014-08-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Conformational Analysis of the Streptococcus Pneumoniae Hyaluronate Lyase and Characterization of its Hyaluronan-Specific Carbohydrate-Binding Module.
J.Biol.Chem., 289, 2014
|
|
4D5P
 
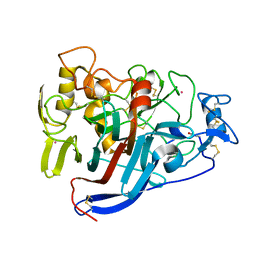 | | Hypocrea jecorina cellobiohydrolase Cel7A E217Q soaked with xylopentaose. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLULOSE 1,4-BETA-CELLOBIOSIDASE, COBALT (II) ION, ... | | Authors: | Momeni, M.H, Ubhayasekera, W, Stahlberg, J, Hansson, H. | | Deposit date: | 2014-11-07 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural Insights Into the Inhibition of Cellobiohydrolase Cel7A by Xylooligosaccharides.
FEBS J., 282, 2015
|
|
4IBJ
 
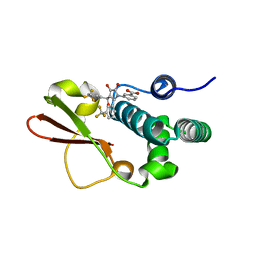 | | Ebola virus VP35 bound to small molecule | | Descriptor: | 3-{(5S)-3-hydroxy-2-oxo-4-[3-(trifluoromethyl)benzoyl]-5-[3-(trifluoromethyl)phenyl]-2,5-dihydro-1H-pyrrol-1-yl}benzoic acid, Polymerase cofactor VP35 | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K. | | Deposit date: | 2012-12-08 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
4D5J
 
 | | Hypocrea jecorina cellobiohydrolase Cel7A E217Q soaked with xylotriose. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLULOSE 1,4-BETA-CELLOBIOSIDASE, COBALT (II) ION, ... | | Authors: | Momeni, M.H, Ubhayasekera, W, Stahlberg, J, Hansson, H. | | Deposit date: | 2014-11-05 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Insights Into the Inhibition of Cellobiohydrolase Cel7A by Xylooligosaccharides.
FEBS J., 282, 2015
|
|
8WD2
 
 | | The Crystal Structure of p53 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, PHOSPHATE ION, ... | | Authors: | Wang, F, Cheng, W, Yuan, Z, Qi, J, Lu, Y. | | Deposit date: | 2023-09-14 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Crystal Structure of p53 from Biortus.
To Be Published
|
|
8WF7
 
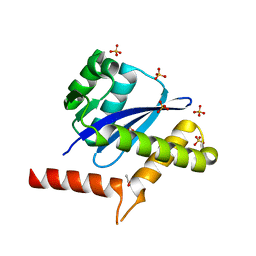 | | The Crystal Structure of integrase from Biortus | | Descriptor: | ACETATE ION, Integrase, SULFATE ION | | Authors: | Wang, F, Cheng, W, Yuan, Z, Qi, J, Li, J. | | Deposit date: | 2023-09-19 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Crystal Structure of integrase from Biortus
To Be Published
|
|
8WWX
 
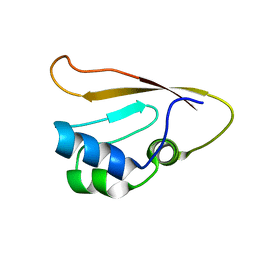 | |
4IGV
 
 | | Crystal structure of kirola (Act d 11) | | Descriptor: | CHLORIDE ION, Kirola, UNKNOWN LIGAND | | Authors: | Chruszcz, M, Ciardiello, M.A, Giangrieco, I, Osinski, T, Minor, W. | | Deposit date: | 2012-12-18 | | Release date: | 2013-09-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and bioinformatic analysis of the kiwifruit allergen Act d 11, a member of the family of ripening-related proteins.
Mol.Immunol., 56, 2013
|
|
8WRB
 
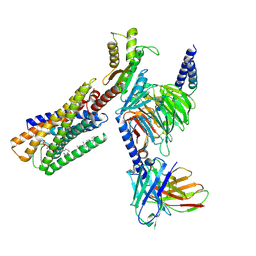 | | Lysophosphatidylserine receptor GPR34-Gi complex | | Descriptor: | Antibody fragment scFv16, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Gong, W, Liu, G, Li, X, Wang, Y, Zhang, X. | | Deposit date: | 2023-10-13 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural basis for ligand recognition and signaling of the lysophosphatidylserine receptors GPR34 and GPR174.
Plos Biol., 21, 2023
|
|
8WR8
 
 | |
4CRF
 
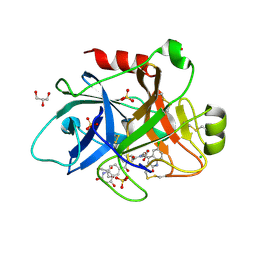 | | Creating novel F1 inhibitors through fragment based lead generation and structure aided drug design | | Descriptor: | CHLORIDE ION, Coagulation factor XIa light chain, GLYCEROL, ... | | Authors: | Sandmark, J, Oster, L, Fjellstrom, O, Akkaya, S, Beisel, H.G, Eriksson, P.O, Erixon, K, Gustafsson, D, Jurva, U, Kang, D, Karis, D, Knecht, W, Nerme, V, Nilsson, I, Olsson, T, Redzic, A, Roth, R, Tigerstrom, A. | | Deposit date: | 2014-02-26 | | Release date: | 2015-02-11 | | Last modified: | 2022-05-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Creating Novel Activated Factor Xi Inhibitors Through Fragment Based Lead Generation and Structure Aided Drug Design.
Plos One, 10, 2015
|
|
4IHR
 
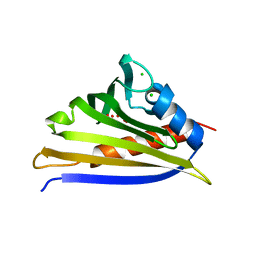 | | Crystal structure of recombinant kirola (Act d 11) | | Descriptor: | CHLORIDE ION, Kirola, UNKNOWN LIGAND | | Authors: | Osinski, T, Majorek, K.A, Ciardiello, M.A, Chruszcz, M, Minor, W. | | Deposit date: | 2012-12-19 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and bioinformatic analysis of the kiwifruit allergen Act d 11, a member of the family of ripening-related proteins.
Mol.Immunol., 56, 2013
|
|
4IBI
 
 | | Ebola virus VP35 bound to small molecule | | Descriptor: | 3-{(2S)-2-(7-chloro-1,3-benzodioxol-5-yl)-4-hydroxy-5-oxo-3-[3-(trifluoromethyl)benzoyl]-2,5-dihydro-1H-pyrrol-1-yl}benzoic acid, Polymerase cofactor VP35 | | Authors: | Brown, C.S, Leung, D.W, Xu, W, Borek, D.M, Otwinowski, Z, Ramanan, P, Stubbs, A.J, Peterson, D.S, Binning, J.M, Amarasinghe, G.K. | | Deposit date: | 2012-12-08 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.473 Å) | | Cite: | In Silico Derived Small Molecules Bind the Filovirus VP35 Protein and Inhibit Its Polymerase Cofactor Activity.
J.Mol.Biol., 426, 2014
|
|
8WTF
 
 | | The Crystal Structure of IRAK4 from Biortus | | Descriptor: | 1,2-ETHANEDIOL, 1-{[(2S,3S,4S)-3-ethyl-4-fluoro-5-oxopyrrolidin-2-yl]methoxy}-7-methoxyisoquinoline-6-carboxamide, Interleukin-1 receptor-associated kinase 4, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Xu, Y. | | Deposit date: | 2023-10-18 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of IRAK4 from Biortus
To Be Published
|
|
8WR5
 
 | | The Crystal Structure of Mms2 from Biortus | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Ubiquitin-conjugating enzyme E2 variant 2 | | Authors: | Wang, F, Cheng, W, Yuan, Z, Lin, D, Guo, S. | | Deposit date: | 2023-10-13 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of Mms2 from Biortus.
To Be Published
|
|
