6BM4
 
 | | Pol II elongation complex with an abasic lesion at i-1 position,soaking UMPNPP | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, DNA (5'-D(P*CP*AP*(3DR)P*CP*TP*CP*TP*TP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, W, Wang, D. | | Deposit date: | 2017-11-13 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.951 Å) | | Cite: | Structural basis of transcriptional stalling and bypass of abasic DNA lesion by RNA polymerase II.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8IK2
 
 | | RhlA exhibits dual thioesterase and acyltransferase activities during rhamnolipid biosynthesis | | Descriptor: | (3~{S})-3-oxidanyldecanoic acid, 3-(3-hydroxydecanoyloxy)decanoate synthase | | Authors: | Tang, T, Fu, L.H, Xie, W.H, Luo, Y.Z, Zhang, Y.T, Si, T. | | Deposit date: | 2023-02-28 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | RhlA Exhibits Dual Thioesterase and Acyltransferase Activities during Rhamnolipid Biosynthesis
Acs Catalysis, 13, 2023
|
|
6K7W
 
 | | Solution Structure of the CS1 Domain of USP19 | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 19 | | Authors: | Xue, W, Hu, H.Y. | | Deposit date: | 2019-06-10 | | Release date: | 2020-06-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Domain interactions reveal auto-inhibition of the deubiquitinating enzyme USP19 and its activation by HSP90 in the modulation of huntingtin aggregation.
Biochem.J., 477, 2020
|
|
1FK1
 
 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH LAURIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | Descriptor: | FORMIC ACID, LAURIC ACID, NON-SPECIFIC LIPID TRANSFER PROTEIN | | Authors: | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 2000-08-09 | | Release date: | 2001-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
1FK6
 
 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH ALPHA-LINOLENIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | Descriptor: | ALPHA-LINOLENIC ACID, FORMIC ACID, NON-SPECIFIC LIPID TRANSFER PROTEIN | | Authors: | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 2000-08-09 | | Release date: | 2001-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
8S9U
 
 | |
1FKH
 
 | | DESIGN, SYNTHESIS, AND KINETIC EVALUATION OF HIGH-AFFINITY FKBP LIGANDS, AND THE X-RAY CRYSTAL STRUCTURES OF THEIR COMPLEXES WITH FKBP12 | | Descriptor: | 1-CYCLOHEXYL-3-PHENYL-1-PROPYL-1-(3,3-DIMETHYL-1,2-DIOXYPENTYL)-2-PIPERIDINE CARBOXYLATE, FK506 BINDING PROTEIN | | Authors: | Holt, D.A, Luengo, J.I, Yamashita, D.S, Oh, H.-J, Konialian, A.L, Yen, H.-K, Rozamus, L.W, Brandt, M, Bossard, M.J, Levy, M.A, Eggleston, D.S, Stout, T.J, Liang, J, Schultz, L.W, Clardy, J. | | Deposit date: | 1993-08-05 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | DESIGN, SYNTHESIS, AND KINETIC EVALUATION OF HIGH-AFFINITY FKBP LIGANDS AND THE X-RAY CRYSTAL-STRUCTURES OF THEIR COMPLEXES WITH FKBP12.
J.Am.Chem.Soc., 115, 1993
|
|
8S9T
 
 | | CRISPR-Cas type III-D effector complex | | Descriptor: | CRISPR RNA, Cas10, Cas7-2x, ... | | Authors: | Schwartz, E.A, Taylor, D.W. | | Deposit date: | 2023-03-30 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | RNA targeting and cleavage by the type III-Dv CRISPR effector complex.
Nat Commun, 15, 2024
|
|
3MR3
 
 | | Human DNA polymerase eta - DNA ternary complex with the 3'T of a CPD in the active site (TT1) | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(*CP*AP*(TTD)P*AP*TP*GP*AP*CP*GP*CP*T)-3'), DNA (5'-D(*CP*AP*GP*CP*GP*TP*CP*AP*T)-3'), ... | | Authors: | Biertumpfel, C, Zhao, Y, Ramon-Maiques, S, Gregory, M.T, Lee, J.Y, Yang, W. | | Deposit date: | 2010-04-28 | | Release date: | 2010-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and mechanism of human DNA polymerase eta.
Nature, 465, 2010
|
|
4P9A
 
 | | X-ray Crystal Structure of PA protein from Influenza strain H7N9 | | Descriptor: | GLYCEROL, Polymerase PA | | Authors: | Fairman, J.W, Moen, S.O, Clifton, M.C, Edwards, T.E, Lorimer, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2014-04-02 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1999 Å) | | Cite: | Structural analysis of H1N1 and H7N9 influenza A virus PA in the absence of PB1.
Sci Rep, 4, 2014
|
|
7UNY
 
 | | Crystal structure of D2 nanobody in complex with PfCSS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cysteine-rich small secreted protein CSS, ... | | Authors: | Scally, S.W, Lim, P.S, Cowman, A.F. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.13 Å) | | Cite: | PCRCR complex is essential for invasion of human erythrocytes by Plasmodium falciparum.
Nat Microbiol, 7, 2022
|
|
3MR5
 
 | | Human DNA polymerase eta - DNA ternary complex with a CPD 1bp upstream of the active site (TT3) | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*AP*CP*GP*TP*CP*AP*TP*AP*A)-3'), DNA (5'-D(*TP*AP*AP*CP*(TTD)P*AP*TP*GP*AP*CP*GP*A)-3'), ... | | Authors: | Biertumpfel, C, Zhao, Y, Ramon-Maiques, S, Gregory, M.T, Lee, J.Y, Yang, W. | | Deposit date: | 2010-04-28 | | Release date: | 2010-06-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and mechanism of human DNA polymerase eta.
Nature, 465, 2010
|
|
8R33
 
 | |
4AB2
 
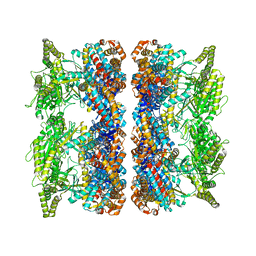 | | ATP-triggered molecular mechanics of the chaperonin GroEL | | Descriptor: | 60 KDA CHAPERONIN, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Clare, D.K, Vasishtan, D, Stagg, S, Quispe, J, Farr, G.W, Topf, M, Horwich, A.L, Saibil, H.R. | | Deposit date: | 2011-12-06 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | ATP-Triggered Conformational Changes Delineate Substrate-Binding and -Folding Mechanics of the Groel Chaperonin.
Cell(Cambridge,Mass.), 149, 2012
|
|
8QYG
 
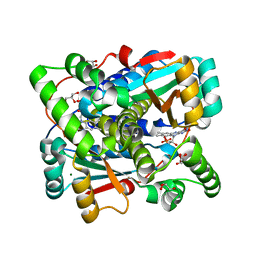 | | Crystal structure of Nitroreductase from Bacillus tequilensis | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Rozeboom, H.J, Russo, S, Fraaije, M.W, Poelarends, G.J. | | Deposit date: | 2023-10-26 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Biochemical, kinetic, and structural characterization of a Bacillus tequilensis nitroreductase.
Febs J., 291, 2024
|
|
7U8Z
 
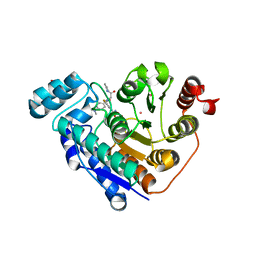 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 2 (CD2) complexed with fluorinated peptoid inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-({N-[2-(benzylamino)-2-oxoethyl]-4-(dimethylamino)benzamido}methyl)-3-fluoro-N-hydroxybenzamide, ACETATE ION, ... | | Authors: | Watson, P.R, Cragin, A.D, Christianson, D.W. | | Deposit date: | 2022-03-09 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Development of Fluorinated Peptoid-Based Histone Deacetylase (HDAC) Inhibitors for Therapy-Resistant Acute Leukemia.
J.Med.Chem., 65, 2022
|
|
3T7N
 
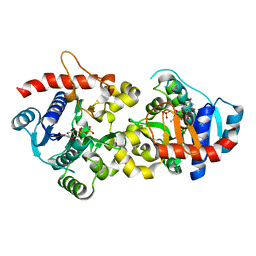 | | Crystal Structure of Human Glycogenin-1 (GYG1) complexed with manganese and UDP, in a monoclinic closed form | | Descriptor: | Glycogenin-1, MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE | | Authors: | Chaikuad, A, Froese, D.S, Krysztofinska, E, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Oppermann, U, Yue, W.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-07-30 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Conformational plasticity of glycogenin and its maltosaccharide substrate during glycogen biogenesis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3M12
 
 | |
6CCR
 
 | | Selenomethionyl derivative of a GID4 fragment | | Descriptor: | Glucose-induced degradation protein 4 homolog, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-07 | | Release date: | 2018-04-04 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis of GID4-mediated recognition of degrons for the Pro/N-end rule pathway.
Nat. Chem. Biol., 14, 2018
|
|
2VMB
 
 | | The three-dimensional structure of the cytoplasmic domains of EpsF from the Type 2 Secretion System of Vibrio cholerae | | Descriptor: | CALCIUM ION, GENERAL SECRETION PATHWAY PROTEIN F | | Authors: | Abendroth, J, Korotkov, K.V, Mitchell, D.D, Kreger, A, Hol, W.G.J. | | Deposit date: | 2008-01-25 | | Release date: | 2009-02-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Three-Dimensional Structure of the Cytoplasmic Domains of Epsf from the Type 2 Secretion System of Vibrio Cholerae.
J.Struct.Biol., 166, 2009
|
|
3TGH
 
 | | GAP50 the anchor in the inner membrane complex of Plasmodium | | Descriptor: | COBALT (II) ION, DIMETHYL SULFOXIDE, Glideosome-associated protein 50, ... | | Authors: | Bosch, J, Paige, M.H, Vaidya, A, Bergman, L, Hol, W.G.J. | | Deposit date: | 2011-08-17 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of GAP50, the anchor of the invasion machinery in the inner membrane complex of Plasmodium falciparum.
J.Struct.Biol., 178, 2012
|
|
4CQX
 
 | | H5 (tyTy) Del133/Ile155Thr Mutant Haemagglutinin in Complex with Human Receptor Analogue 6'SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HAEMAGGLUTININ HA1, HAEMAGGLUTININ HA2, ... | | Authors: | Xiong, X, Xiao, H, Martin, S.R, Coombs, P.J, Liu, J, Collins, P.J, Vachieri, S.G, Walker, P.A, Lin, Y.P, McCauley, J.W, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2014-02-21 | | Release date: | 2014-05-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enhanced Human Receptor Binding by H5 Haemagglutinins.
Virology, 456, 2014
|
|
2O36
 
 | |
8G17
 
 | | CryoEM structure of wild-type GAPDH | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Choi, W.Y, Wu, H, Cheng, Y.F. | | Deposit date: | 2023-02-01 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (1.98 Å) | | Cite: | Efficient tagging of endogenous proteins in human cell lines for structural studies by single-particle cryo-EM.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1FJG
 
 | | STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH THE ANTIBIOTICS STREPTOMYCIN, SPECTINOMYCIN, AND PAROMOMYCIN | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Carter, A.P, Clemons Jr, W.M, Brodersen, D.E, Wimberly, B.T, Morgan-Warren, R.J, Ramakrishnan, V. | | Deposit date: | 2000-08-08 | | Release date: | 2000-09-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics
Nature, 407, 2000
|
|
