5L92
 
 | |
1REW
 
 | | Structural refinement of the complex of bone morphogenetic protein 2 and its type IA receptor | | Descriptor: | bone morphogenetic protein 2, bone morphogenetic protein receptor type IA | | Authors: | Keller, S, Nickel, J, Zhang, J.-L, Sebald, W, Mueller, T.D. | | Deposit date: | 2003-11-07 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.863 Å) | | Cite: | Molecular recognition of BMP-2 and BMP receptor IA.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1RFS
 
 | | RIESKE SOLUBLE FRAGMENT FROM SPINACH | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, RIESKE PROTEIN | | Authors: | Carrell, C.J, Zhang, H, Cramer, W.A, Smith, J.L. | | Deposit date: | 1997-08-14 | | Release date: | 1998-01-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Biological identity and diversity in photosynthesis and respiration: structure of the lumen-side domain of the chloroplast Rieske protein.
Structure, 5, 1997
|
|
7C31
 
 | | Crystal structure of the grapevine defensin VvK1 | | Descriptor: | Knot1 domain-containing protein | | Authors: | Chen, M.W, Chang, S.C, Chandy, K.G, Luo, D. | | Deposit date: | 2020-05-10 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Modulation of Lymphocyte Potassium Channel KV1.3 by Membrane-Penetrating, Joint-Targeting Immunomodulatory Plant Defensin.
Acs Pharmacol Transl Sci, 3, 2020
|
|
1ROP
 
 | |
5L90
 
 | |
4V2F
 
 | | Tetracycline repressor TetR(D), unliganded | | Descriptor: | TETRACYCLINE REPRESSOR PROTEIN CLASS D | | Authors: | Werten, S, Orth, P, Saenger, W, Hinrichs, W. | | Deposit date: | 2014-10-09 | | Release date: | 2014-12-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Tetracycline Repressor Allostery Does not Depend on Divalent Metal Recognition.
Biochemistry, 53, 2014
|
|
4URF
 
 | | Molecular Genetic and Crystal Structural Analysis of 1-(4- Hydroxyphenyl)-Ethanol Dehydrogenase from Aromatoleum aromaticum EbN1 | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, ACETATE ION, BICARBONATE ION, ... | | Authors: | Buesing, I, Hoeffken, H.W, Breuer, M, Woehlbrand, L, Hauer, B, Rabus, R. | | Deposit date: | 2014-06-28 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Molecular Genetic and Crystal Structural Analysis of 1-(4-Hydroxyphenyl)-Ethanol Dehydrogenase from 'Aromatoleum Aromaticum' Ebn1.
J.Mol.Microbiol.Biotechnol., 25, 2015
|
|
1R9Q
 
 | | structure analysis of ProX in complex with proline betaine | | Descriptor: | 1,1-DIMETHYL-PROLINIUM, Glycine betaine-binding periplasmic protein, UNKNOWN ATOM OR ION | | Authors: | Schiefner, A, Breed, J, Bosser, L, Kneip, S, Gade, J, Holtmann, G, Diederichs, K, Welte, W, Bremer, E. | | Deposit date: | 2003-10-30 | | Release date: | 2004-02-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Cation-pi Interactions as Determinants for Binding of the Compatible Solutes Glycine Betaine and Proline Betaine by the Periplasmic Ligand-binding Protein ProX from Escherichia coli
J.BIOL.CHEM., 279, 2004
|
|
4V2G
 
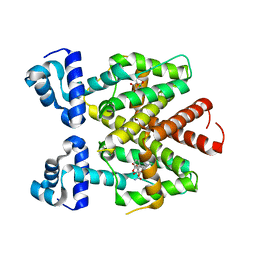 | | Tetracycline repressor TetR(D) bound to chlortetracycline and iso- chlortetracycline | | Descriptor: | 7-CHLOROTETRACYCLINE, ISO-7-CHLORTETRACYCLINE, MAGNESIUM ION, ... | | Authors: | Werten, S, Orth, P, Saenger, W, Hinrichs, W. | | Deposit date: | 2014-10-09 | | Release date: | 2014-12-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Tetracycline Repressor Allostery Does not Depend on Divalent Metal Recognition.
Biochemistry, 53, 2014
|
|
5LCZ
 
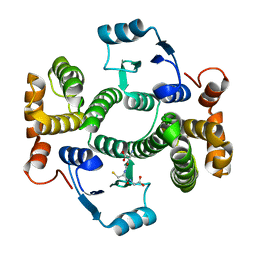 | | Chimeric GST | | Descriptor: | GLUTATHIONE, Glutathione S-transferase A1,Glutathione S-transferase alpha-2,Glutathione S-transferase A1,Glutathione S-transferase alpha-2,Glutathione S-transferase A1 | | Authors: | Axarli, A, Muleta, A.W, Chronopoulou, E.G, Papageorgiou, A.C, Labrou, N.E. | | Deposit date: | 2016-06-23 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.325 Å) | | Cite: | Directed evolution of glutathione transferases towards a selective glutathione-binding site and improved oxidative stability.
Biochim. Biophys. Acta, 1861, 2017
|
|
1RTG
 
 | |
1RJA
 
 | |
1RL6
 
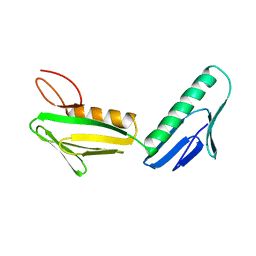 | | RIBOSOMAL PROTEIN L6 | | Descriptor: | PROTEIN (RIBOSOMAL PROTEIN L6) | | Authors: | Golden, B.L, Davies, C, Ramakrishnan, V, White, S.W. | | Deposit date: | 1999-01-14 | | Release date: | 1999-02-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ribosomal protein L6: structural evidence of gene duplication from a primitive RNA binding protein.
EMBO J., 12, 1993
|
|
5LHA
 
 | |
7C2E
 
 | | GLP-1R-Gs complex structure with a small molecule full agonist | | Descriptor: | 2-[[4-[6-[(4-cyano-2-fluoranyl-phenyl)methoxy]pyridin-2-yl]-3,6-dihydro-2~{H}-pyridin-1-yl]methyl]-3-[[(2~{S})-oxetan-2-yl]methyl]imidazo[4,5-b]pyridine-5-carboxylic acid, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ma, H, Yuan, D.P, Huang, W, Wenge, Z, Xu, E. | | Deposit date: | 2020-05-07 | | Release date: | 2020-08-26 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural insights into the activation of GLP-1R by a small molecule agonist.
Cell Res., 30, 2020
|
|
1RS6
 
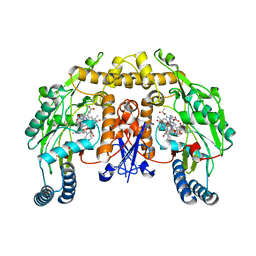 | | Rat neuronal NOS heme domain with D-lysine-D-nitroarginine amide bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, D-MANNITOL, ... | | Authors: | Flinspach, M, Li, H, Jamal, J, Yang, W, Huang, H, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2003-12-09 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the Neuronal and Endothelial Nitric Oxide Synthase Heme Domain with d-Nitroarginine-Containing Dipeptide Inhibitors Bound.
Biochemistry, 43, 2004
|
|
1R8G
 
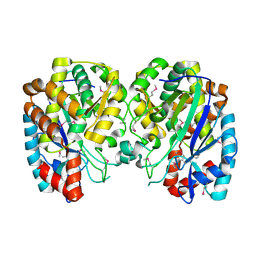 | | Structure and function of YbdK | | Descriptor: | Hypothetical protein ybdK | | Authors: | Lehmann, C, Doseeva, V, Pullalarevu, S, Krajewski, W, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2003-10-24 | | Release date: | 2004-08-17 | | Last modified: | 2021-07-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | YbdK is a carboxylate-amine ligase with a gamma-glutamyl:Cysteine ligase activity: crystal structure and enzymatic assays
PROTEINS: STRUCT.,FUNCT.,GENET., 56, 2004
|
|
1R88
 
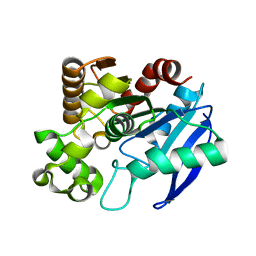 | | The crystal structure of Mycobacterium tuberculosis MPT51 (FbpC1) | | Descriptor: | MPT51/MPB51 antigen | | Authors: | Wilson, R.A, Maughan, W.N, Kremer, L, Besra, G.S, Futterer, K. | | Deposit date: | 2003-10-23 | | Release date: | 2003-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | The structure of Mycobacterium tuberculosis MPT51 (FbpC1) defines a new family of non-catalytic alpha/beta hydrolases.
J.Mol.Biol., 335, 2004
|
|
1R9L
 
 | | structure analysis of ProX in complex with glycine betaine | | Descriptor: | Glycine betaine-binding periplasmic protein, TRIMETHYL GLYCINE, UNKNOWN ATOM OR ION | | Authors: | Schiefner, A, Breed, J, Bosser, L, Kneip, S, Gade, J, Holtmann, G, Diederichs, K, Welte, W, Bremer, E. | | Deposit date: | 2003-10-30 | | Release date: | 2004-02-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Cation-pi Interactions as Determinants for Binding of the Compatible Solutes Glycine Betaine and Proline Betaine by the Periplasmic Ligand-binding Protein ProX from Escherichia coli
J.BIOL.CHEM., 279, 2004
|
|
5LH9
 
 | |
4V94
 
 | | Molecular architecture of the eukaryotic chaperonin TRiC/CCT derived by a combination of chemical crosslinking and mass-spectrometry, XL-MS | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Leitner, A, Joachimiak, L.A, Bracher, A, Walzthoeni, T, Chen, B, Monkemeyer, L, Pechmann, S, Holmes, S, Cong, Y, Ma, B, Ludtke, S, Chiu, W, Hartl, F.U, Aebersold, R, Frydman, J. | | Deposit date: | 2012-01-11 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The Molecular Architecture of the Eukaryotic Chaperonin TRiC/CCT.
Structure, 20, 2012
|
|
4V6E
 
 | | Crystal structure of the E. coli 70S ribosome in an intermediate state of ratcheting | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, W, Dunkle, J.A, Cate, J.H.D. | | Deposit date: | 2009-06-28 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (3.712 Å) | | Cite: | Structures of the ribosome in intermediate States of ratcheting.
Science, 325, 2009
|
|
4V55
 
 | | Crystal structure of the bacterial ribosome from Escherichia coli in complex with gentamicin and ribosome recycling factor (RRF). | | Descriptor: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, 16S rRNA, 23S rRNA, ... | | Authors: | Borovinskaya, M.A, Pai, R.D, Zhang, W, Schuwirth, B.-S, Holton, J.M, Hirokawa, G, Kaji, H, Kaji, A, Cate, J.H.D. | | Deposit date: | 2007-06-17 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural basis for aminoglycoside inhibition of bacterial ribosome recycling.
Nat.Struct.Mol.Biol., 14, 2007
|
|
1R5L
 
 | | Crystal Structure of Human Alpha-Tocopherol Transfer Protein Bound to its Ligand | | Descriptor: | (2R)-2,5,7,8-TETRAMETHYL-2-[(4R,8R)-4,8,12-TRIMETHYLTRIDECYL]CHROMAN-6-OL, PROTEIN (Alpha-tocopherol transfer protein) | | Authors: | Min, K.C, Kovall, R.A, Hendrickson, W.A. | | Deposit date: | 2003-10-10 | | Release date: | 2003-11-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of human alpha-tocopherol transfer protein bound to its ligand: Implications for ataxia with vitamin E deficiency
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
