3V4D
 
 | | Crystal structure of RutC protein a member of the YjgF family from E.coli | | Descriptor: | Aminoacrylate peracid reductase RutC | | Authors: | Knapik, A.A, Petkowski, J.J, Otwinowski, Z, Cymborowski, M.T, Cooper, D.R, Chruszcz, M, Porebski, P.J, Niedzialkowska, E, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-14 | | Release date: | 2012-01-04 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of Escherichia coli RutC, a member of the YjgF family and putative aminoacrylate peracid reductase of the rut operon.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
1P46
 
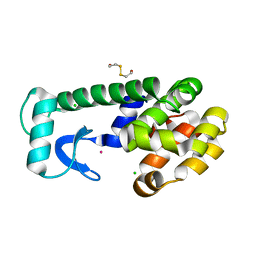 | | T4 lysozyme core repacking mutant M106I/TA | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME, ... | | Authors: | Mooers, B.H, Datta, D, Baase, W.A, Zollars, E.S, Mayo, S.L, Matthews, B.W. | | Deposit date: | 2003-04-21 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Repacking the Core of T4 lysozyme by automated design
J.Mol.Biol., 332, 2003
|
|
6MVE
 
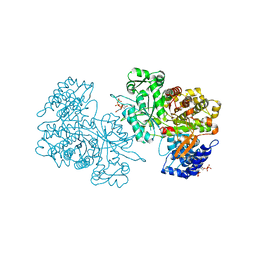 | | Reduced X-ray crystal structure of Bacillus subtilis ribonucleotide reductase NrdE alpha subunit with TTP, ATP, and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Thomas, W.C, Brooks, F.P, Bacik, J.P, Ando, N. | | Deposit date: | 2018-10-25 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Convergent allostery in ribonucleotide reductase.
Nat Commun, 10, 2019
|
|
1U7J
 
 | | Solution structure of a diiron protein model | | Descriptor: | Four-helix bundle model, ZINC ION | | Authors: | Maglio, O, Nastri, F, Calhoun, J.R, Lahr, S, Pavone, V, DeGrado, W.F, Lombardi, A. | | Deposit date: | 2004-08-04 | | Release date: | 2005-03-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Analysis and Design of Turns in alpha-Helical Hairpins
J.Mol.Biol., 346, 2005
|
|
6MKI
 
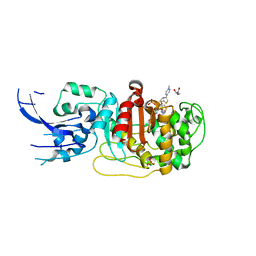 | | Crystal structure of penicillin-binding protein 4 (PBP4) from Enterococcus faecalis in the ceftaroline-bound form | | Descriptor: | Ceftaroline, bound form, GLYCEROL, ... | | Authors: | D'Andrea, E.D, Moon, T.M, Peti, W, Page, R. | | Deposit date: | 2018-09-25 | | Release date: | 2018-10-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.984 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
3UYL
 
 | | Spinosyn Rhamnosyltransferase SpnG complexed with thymidine diphosphate | | Descriptor: | MAGNESIUM ION, NDP-rhamnosyltransferase, THYMIDINE-5'-DIPHOSPHATE, ... | | Authors: | Isiorho, E.A, Liu, H.-W, Keatinge-Clay, A.T. | | Deposit date: | 2011-12-06 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Studies of the Spinosyn Rhamnosyltransferase, SpnG.
Biochemistry, 51, 2012
|
|
5MTP
 
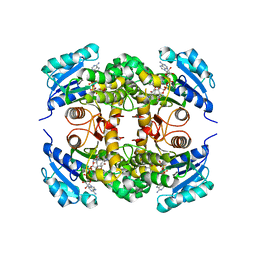 | | Crystal structure of M. tuberculosis InhA inhibited by PT514 | | Descriptor: | 2-(2-methylphenoxy)-5-[(4-phenyl-1H-1,2,3-triazol-1-yl)methyl]phenol, CHLORIDE ION, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Eltschkner, S, Pschibul, A, Spagnuolo, L.A, Yu, W, Tonge, P.J, Kisker, C. | | Deposit date: | 2017-01-10 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evaluating the Contribution of Transition-State Destabilization to Changes in the Residence Time of Triazole-Based InhA Inhibitors.
J. Am. Chem. Soc., 139, 2017
|
|
1PWZ
 
 | | Crystal structure of the haloalcohol dehalogenase HheC complexed with (R)-styrene oxide and chloride | | Descriptor: | CHLORIDE ION, R-STYRENE OXIDE, halohydrin dehalogenase | | Authors: | de Jong, R.M, Tiesinga, J.J.W, Rozeboom, H.J, Kalk, K.H, Tang, L, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2003-07-02 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Mechanism of a Bacterial Haloalcohol Dehalogenase: a new variation of the short-chain dehydrogenase/reductase fold without an NAD(P)H binding site
EMBO J., 22, 2003
|
|
6MKJ
 
 | | Crystal structure of penicillin binding protein 5 (PBP5) from Enterococcus faecium in the closed conformation | | Descriptor: | penicillin binding protein 5 (PBP5) | | Authors: | Moon, T.M, Soares, A, D'Andrea, E.D, Jaconcic, J, Peti, W, Page, R. | | Deposit date: | 2018-09-25 | | Release date: | 2018-10-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.864 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
4G5S
 
 | | Structure of LGN GL3/Galphai3 complex | | Descriptor: | CITRIC ACID, G-protein-signaling modulator 2, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Jia, M, Li, J, Zhu, J, Wen, W, Zhang, M, Wang, W. | | Deposit date: | 2012-07-18 | | Release date: | 2012-09-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Crystal Structures of the scaffolding protein LGN reveal the general mechanism by which GoLoco binding motifs inhibit the release of GDP from Galphai subunits in G-coupled heterotrimeric proteins
To be Published
|
|
6I5E
 
 | | X-ray structure of apo human soluble Epoxide Hydrolase C-terminal Domain (hsEH CTD) | | Descriptor: | Bifunctional epoxide hydrolase 2 | | Authors: | Abis, G, Kopec, J, Yue, W.W, Conte, M.R. | | Deposit date: | 2018-11-13 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 15-deoxy-Delta12,14-Prostaglandin J2inhibits human soluble epoxide hydrolase by a dual orthosteric and allosteric mechanism.
Commun Biol, 2, 2019
|
|
4G73
 
 | | Crystal structure of NDH with NADH and Quinone | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2,3-DIMETHOXY-5-METHYL-6-(3,11,15,19-TETRAMETHYL-EICOSA-2,6,10,14,18-PENTAENYL)-[1,4]BENZOQUINONE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Li, W, Feng, Y, Ge, J, Yang, M. | | Deposit date: | 2012-07-19 | | Release date: | 2012-10-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.522 Å) | | Cite: | Structural insight into the type-II mitochondrial NADH dehydrogenases.
Nature, 491, 2012
|
|
6O0R
 
 | | Crystal structure of the TIR domain from human SARM1 in complex with glycerol | | Descriptor: | GLYCEROL, Sterile alpha and TIR motif-containing protein 1 | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Deerain, N, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
6I4X
 
 | | Crystal structure of SOCS2:Elongin C:Elongin B in complex with erythropoietin receptor peptide | | Descriptor: | DI(HYDROXYETHYL)ETHER, Elongin-B, Elongin-C, ... | | Authors: | Kung, W.W, Ramachandran, S, Makukhin, N, Bruno, E, Ciulli, A. | | Deposit date: | 2018-11-12 | | Release date: | 2019-05-29 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural insights into substrate recognition by the SOCS2 E3 ubiquitin ligase.
Nat Commun, 10, 2019
|
|
6O0U
 
 | | Crystal structure of the TIR domain H685A mutant from human SARM1 | | Descriptor: | Sterile alpha and TIR motif-containing protein 1 | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
6O23
 
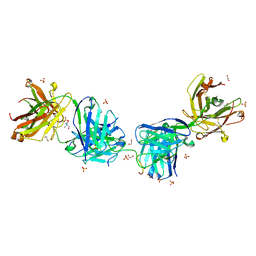 | | Crystal structure of 2243 Fab in complex with circumsporozoite protein NANP5 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2243 Fab heavy chain, ... | | Authors: | Scally, S.W, Bosch, A, Prieto, K, Murugan, R, Wardemann, H, Julien, J.P. | | Deposit date: | 2019-02-22 | | Release date: | 2020-03-04 | | Last modified: | 2020-09-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolution of protective human antibodies against Plasmodium falciparum circumsporozoite protein repeat motifs.
Nat. Med., 26, 2020
|
|
1PHO
 
 | |
6NYL
 
 | | Helicobacter pylori Vacuolating Cytotoxin A Oligomeric Assembly 2c (OA-2c) | | Descriptor: | Vacuolating cytotoxin autotransporter | | Authors: | Zhang, K, Zhang, H, Li, S, Au, S, Chiu, W. | | Deposit date: | 2019-02-11 | | Release date: | 2019-03-27 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures ofHelicobacter pylorivacuolating cytotoxin A oligomeric assemblies at near-atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6O0M
 
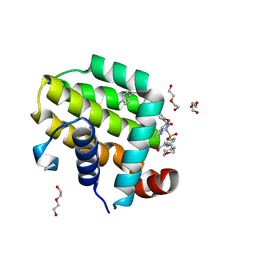 | | crystal structure of BCL-2 F104L mutation with venetoclax | | Descriptor: | 4-{4-[(4'-chloro-5,5-dimethyl[3,4,5,6-tetrahydro[1,1'-biphenyl]]-2-yl)methyl]piperazin-1-yl}-N-[(3-nitro-4-{[(oxan-4-yl )methyl]amino}phenyl)sulfonyl]-2-[(1H-pyrrolo[2,3-b]pyridin-5-yl)oxy]benzamide, Apoptosis regulator Bcl-2,Bcl-2-like protein 1,Apoptosis regulator Bcl-2, DI(HYDROXYETHYL)ETHER | | Authors: | Birkinshaw, R.W, Luo, C.S, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-02-16 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of BCL-2 in complex with venetoclax reveal the molecular basis of resistance mutations.
Nat Commun, 10, 2019
|
|
6O0Q
 
 | | Crystal structure of the TIR domain from human SARM1 in complex with ribose | | Descriptor: | CHLORIDE ION, Sterile alpha and TIR motif-containing protein 1, beta-D-ribofuranose | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Deerain, N, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
6NYN
 
 | | Helicobacter pylori Vacuolating Cytotoxin A Oligomeric Assembly 2e (OA-2e) | | Descriptor: | Vacuolating cytotoxin autotransporter | | Authors: | Zhang, K, Zhang, H, Li, S, Au, S, Chiu, W. | | Deposit date: | 2019-02-11 | | Release date: | 2019-03-27 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures ofHelicobacter pylorivacuolating cytotoxin A oligomeric assemblies at near-atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6O0K
 
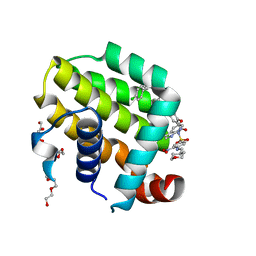 | | crystal structure of BCL-2 with venetoclax | | Descriptor: | 4-{4-[(4'-chloro-5,5-dimethyl[3,4,5,6-tetrahydro[1,1'-biphenyl]]-2-yl)methyl]piperazin-1-yl}-N-[(3-nitro-4-{[(oxan-4-yl )methyl]amino}phenyl)sulfonyl]-2-[(1H-pyrrolo[2,3-b]pyridin-5-yl)oxy]benzamide, Apoptosis regulator Bcl-2, NONAETHYLENE GLYCOL | | Authors: | Birkinshaw, R.W, Luo, C.S, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-02-16 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structures of BCL-2 in complex with venetoclax reveal the molecular basis of resistance mutations.
Nat Commun, 10, 2019
|
|
6O2B
 
 | | Crystal structure of 4493 Fab in complex with circumsporozoite protein DND and anti-kappa VHH domain | | Descriptor: | 1,2-ETHANEDIOL, 4493 Fab heavy chain, 4493 Kappa light chain, ... | | Authors: | Scally, S.W, Bosch, A, Prieto, K, Murugan, R, Wardemann, H, Julien, J.P. | | Deposit date: | 2019-02-22 | | Release date: | 2020-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Evolution of protective human antibodies against Plasmodium falciparum circumsporozoite protein repeat motifs.
Nat. Med., 26, 2020
|
|
6KNT
 
 | | Crystal structure of the metallo-beta-lactamase fold protein YhfI from Bacillus subtilis (space group P4332) | | Descriptor: | Putative metal-dependent hydrolase, ZINC ION | | Authors: | Na, H.W, Namgung, B, Song, W.S, Yoon, S.I. | | Deposit date: | 2019-08-07 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and biochemical analyses of the metallo-beta-lactamase fold protein YhfI from Bacillus subtilis.
Biochem.Biophys.Res.Commun., 519, 2019
|
|
2GZL
 
 | | Structure of 2C-methyl-D-erythritol 2,4-clycodiphosphate synthase complexed with a CDP derived fluorescent inhibitor | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, 5'-O-{[({[2-({[5-(DIMETHYLAMINO)NAPHTHALEN-1-YL]SULFONYL}AMINO)ETHYL]OXY}PHOSPHINATO)OXY]PHOSPHINATO}CYT, GERANYL DIPHOSPHATE, ... | | Authors: | Crane, C.M, Kaiser, J, Ramsden, N.L, Lauw, S, Rohdich, F, Wolfgang, E, Hunter, W.N, Bacher, A, Diederich, F. | | Deposit date: | 2006-05-11 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fluorescent Inhibitors for Ispf, an Enzyme in the Non-Mevalonate Pathway for Isoprenoid Biosynthesis and a Potential Target for Antimalarial Therapy.
Angew.Chem.Int.Ed.Engl., 45, 2006
|
|
