3HYL
 
 | | Crystal Structure of Transketolase from Bacillus anthracis | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Maltseva, N, Kim, Y, Kwon, K, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-06-22 | | Release date: | 2009-06-30 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal Structure of Transketolase from Bacillus anthracis
To be Published
|
|
7U87
 
 | | Product of 13mer primer with activated G monomer diastereomer 1 | | Descriptor: | 5'-O-[(R)-(2-amino-1H-imidazol-1-yl)(sulfanyl)phosphoryl]guanosine, MAGNESIUM ION, RNA (5'-R(*(LCC)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*(G46)P*(G46))-3') | | Authors: | Fang, Z, Szostak, J.W. | | Deposit date: | 2022-03-08 | | Release date: | 2023-03-15 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Catalytic Metal Ion-Substrate Coordination during Nonenzymatic RNA Primer Extension.
J.Am.Chem.Soc., 2024
|
|
6BX6
 
 | | AMP-Activated protein kinase (AMPK) inhibition by SBI-0206965: alpha 2 kinase domain bound to SBI-0206965 | | Descriptor: | 2-({5-bromo-2-[(3,4,5-trimethoxyphenyl)amino]pyrimidin-4-yl}oxy)-N-methylbenzene-1-carboximidic acid, 5'-AMP-activated protein kinase catalytic subunit alpha-2 | | Authors: | Dite, T.A, Langendorf, C.G, Scott, J.W, Oakhill, J.S. | | Deposit date: | 2017-12-17 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | AMP-activated protein kinase selectively inhibited by the type II inhibitor SBI-0206965.
J. Biol. Chem., 293, 2018
|
|
1A31
 
 | | HUMAN RECONSTITUTED DNA TOPOISOMERASE I IN COVALENT COMPLEX WITH A 22 BASE PAIR DNA DUPLEX | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*AP*GP*AP*CP*5IUP*5IU*TP*GP*AP*AP*AP*AP*AP*5IUP*5IUP*5IUP*5IUP*T)-3'), DNA (5'-D(*AP*AP*AP*AP*AP*TP*5IUP*5IUP*5IUP*5IUP*CP*AP*AP*AP*GP*TP*CP*TP*TP*TP*TP*T)-3'), PROTEIN (TOPOISOMERASE I) | | Authors: | Redinbo, M.R, Stewart, L, Kuhn, P, Champoux, J.J, Hol, W.G.J. | | Deposit date: | 1998-01-27 | | Release date: | 1998-08-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA.
Science, 279, 1998
|
|
1A3Q
 
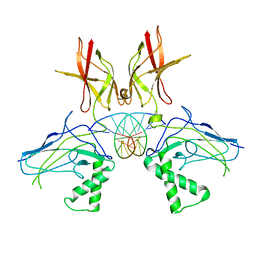 | | HUMAN NF-KAPPA-B P52 BOUND TO DNA | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*AP*AP*TP*CP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*GP*AP*TP*TP*CP*CP*CP*C)-3'), PROTEIN (NUCLEAR FACTOR KAPPA-B P52) | | Authors: | Cramer, P, Larson, C.J, Verdine, G.L, Muller, C.W. | | Deposit date: | 1998-01-23 | | Release date: | 1998-06-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the human NF-kappaB p52 homodimer-DNA complex at 2.1 A resolution.
EMBO J., 16, 1997
|
|
5FKI
 
 | | Pseudorabies virus (PrV) nuclear egress complex proteins fitted as a hexameric lattice into a sub-tomogram average derived from focused- ion beam milled lamellae electron cryo-microscopic data | | Descriptor: | CHLORIDE ION, UL31, UL34 protein, ... | | Authors: | Hagen, C, Dent, K.C, Zeev Ben Mordehai, T, Vasishtan, D, Antonin, W, Mettenleiter, T.C, Gruenewald, K. | | Deposit date: | 2015-10-16 | | Release date: | 2016-03-16 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (35 Å) | | Cite: | Crystal Structure of the Herpesvirus Nuclear Egress Complex Provides Insights Into Inner Nuclear Membrane Remodelling
Cell Rep., 13, 2015
|
|
7U89
 
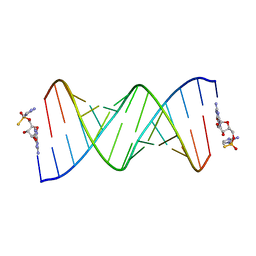 | | Product of 14mer primer with activated G monomer diastereomer 1 | | Descriptor: | 5'-O-[(R)-(2-amino-1H-imidazol-1-yl)(sulfanyl)phosphoryl]guanosine, RNA (5'-R(*(LKC)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*GP*(G46))-3') | | Authors: | Fang, Z, Szostak, J.W. | | Deposit date: | 2022-03-08 | | Release date: | 2023-03-15 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Catalytic Metal Ion-Substrate Coordination during Nonenzymatic RNA Primer Extension.
J.Am.Chem.Soc., 2024
|
|
5FQC
 
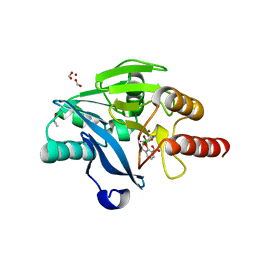 | | Crystal structure of the metallo-beta-lactamase VIM-2 with 2C | | Descriptor: | (4~{R})-4-[[4-(aminomethyl)phenyl]carbonylamino]-3,3-bis(oxidanyl)-2-oxa-3-boranuidabicyclo[4.4.0]deca-1(10),6,8-triene-10-carboxylic acid, BETA-LACTAMASE, DIMETHYL SULFOXIDE, ... | | Authors: | Brem, J, Cain, R, McDonough, M.A, Clifton, I.J, Fishwick, C.W.G, Schofield, C.J. | | Deposit date: | 2015-12-08 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Structural basis of metallo-beta-lactamase, serine-beta-lactamase and penicillin-binding protein inhibition by cyclic boronates.
Nat Commun, 7, 2016
|
|
7U8A
 
 | | Product of 14mer primer with activated G monomer diastereomer 2 | | Descriptor: | 5'-O-[(R)-(2-amino-1H-imidazol-1-yl)(sulfanyl)phosphoryl]guanosine, MAGNESIUM ION, RNA (5'-R(*(LKC)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*GP*(G46))-3') | | Authors: | Fang, Z, Szostak, J.W. | | Deposit date: | 2022-03-08 | | Release date: | 2023-03-15 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Catalytic Metal Ion-Substrate Coordination during Nonenzymatic RNA Primer Extension.
J.Am.Chem.Soc., 2024
|
|
8UCW
 
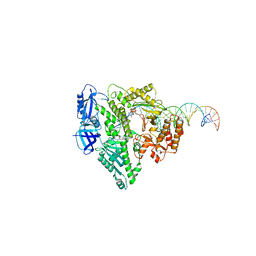 | | Complete DNA termination subcomplex 2 of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA template, ... | | Authors: | Mullins, E.A, Chazin, W.C, Eichman, B.F. | | Deposit date: | 2023-09-27 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8UCV
 
 | | Complete DNA termination subcomplex 1 of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA primase large subunit, ... | | Authors: | Mullins, E.A, Chazin, W.C, Eichman, B.F. | | Deposit date: | 2023-09-27 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
1AAN
 
 | | CRYSTAL STRUCTURE ANALYSIS OF AMICYANIN AND APOAMICYANIN FROM PARACOCCUS DENITRIFICANS AT 2.0 ANGSTROMS AND 1.8 ANGSTROMS RESOLUTION | | Descriptor: | AMICYANIN, COPPER (II) ION | | Authors: | Chen, L, Durley, R.C.E, Lim, L.W, Mathews, F.S. | | Deposit date: | 1992-04-09 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure analysis of amicyanin and apoamicyanin from Paracoccus denitrificans at 2.0 A and 1.8 A resolution.
Protein Sci., 2, 1993
|
|
8UC6
 
 | | Calpain-7:IST1 Complex | | Descriptor: | Calpain-7, IST1 homolog | | Authors: | Paine, E, Whitby, F.G, Hill, C.P, Sunquist, W.I. | | Deposit date: | 2023-09-25 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | The Calpain-7 protease functions together with the ESCRT-III protein IST1 within the midbody to regulate the timing and completion of abscission.
Elife, 12, 2023
|
|
6JI0
 
 | | Structure of RyR2 (F/A/C/Ca2+ dataset) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | Authors: | Gong, D.S, Chi, X.M, Zhou, G.W, Huang, G.X.Y, Lei, J.L, Yan, N. | | Deposit date: | 2019-02-19 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Modulation of cardiac ryanodine receptor 2 by calmodulin.
Nature, 572, 2019
|
|
1A42
 
 | | HUMAN CARBONIC ANHYDRASE II COMPLEXED WITH BRINZOLAMIDE | | Descriptor: | (4R)-2-(2-ethoxyethyl)-4-(ethylamino)-3,4-dihydro-2H-thieno[3,2-e][1,2]thiazine-6-sulfonamide 1,1-dioxide, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Boriack-Sjodin, P.A, Christianson, D.W. | | Deposit date: | 1998-02-10 | | Release date: | 1999-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of murine carbonic anhydrase IV and human carbonic anhydrase II complexed with brinzolamide: molecular basis of isozyme-drug discrimination.
Protein Sci., 7, 1998
|
|
3JWD
 
 | | Structure of HIV-1 gp120 with gp41-Interactive Region: Layered Architecture and Basis of Conformational Mobility | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB 48D HEAVY CHAIN, FAB 48D LIGHT CHAIN, ... | | Authors: | Pancera, M, Majeed, S, Ban, Y.A, Chen, L, Huang, C.C, Kong, L, Kwon, Y.D, Stuckey, J, Zhou, T, Robinson, J.E, Schief, W.R, Sodroski, J, Wyatt, R, Kwong, P.D. | | Deposit date: | 2009-09-18 | | Release date: | 2009-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6BOF
 
 | |
1A5C
 
 | | FRUCTOSE-1,6-BISPHOSPHATE ALDOLASE FROM PLASMODIUM FALCIPARUM | | Descriptor: | FRUCTOSE-1,6-BISPHOSPHATE ALDOLASE | | Authors: | Kim, H, Certa, U, Dobeli, H, Jakob, P, Hol, W.G.J. | | Deposit date: | 1998-02-13 | | Release date: | 1998-06-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of fructose-1,6-bisphosphate aldolase from the human malaria parasite Plasmodium falciparum.
Biochemistry, 37, 1998
|
|
4MVW
 
 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor 1-{3-[(3,5-dichlorobenzyl)amino]propyl}-3-thiophen-3-ylurea (Chem 1433) | | Descriptor: | 1-{3-[(3,5-dichlorobenzyl)amino]propyl}-3-thiophen-3-ylurea, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.Y, Kim, J.E, Wetzel, A.B, de van der Schueren, W.J, Shibata, S, Liu, J, Zhang, Z, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Structures of Trypanosoma brucei Methionyl-tRNA Synthetase with Urea-Based Inhibitors Provide Guidance for Drug Design against Sleeping Sickness.
Plos Negl Trop Dis, 8, 2014
|
|
4ZFL
 
 | |
4MWE
 
 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor 1-(3-{[5-chloro-3-(prop-2-en-1-yl)-2-(prop-2-en-1-yloxy)benzyl]amino}propyl)-3-thiophen-3-ylurea (Chem 1475) | | Descriptor: | 1-(3-{[5-chloro-3-(prop-2-en-1-yl)-2-(prop-2-en-1-yloxy)benzyl]amino}propyl)-3-thiophen-3-ylurea, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.Y, Kim, J.E, Wetzel, A.B, de van der Schueren, W.J, Shibata, S, Liu, J, Zhang, Z, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structures of Trypanosoma brucei Methionyl-tRNA Synthetase with Urea-Based Inhibitors Provide Guidance for Drug Design against Sleeping Sickness.
Plos Negl Trop Dis, 8, 2014
|
|
1AF9
 
 | | TETANUS NEUROTOXIN C FRAGMENT | | Descriptor: | TETANUS NEUROTOXIN | | Authors: | Umland, T.C, Wingert, L, Swaminathan, S, Furey, W.F, Schmidt, J.J, Sax, M. | | Deposit date: | 1997-03-24 | | Release date: | 1998-04-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the receptor binding fragment HC of tetanus neurotoxin.
Nat.Struct.Biol., 4, 1997
|
|
1GG0
 
 | | CRYSTAL STRUCTURE ANALYSIS OF KDOP SYNTHASE AT 3.0 A | | Descriptor: | 3-DEOXY-D-MANNO-OCTULOSONATE 8-PHOSPHATE SYNTHASE, PHOSPHATE ION | | Authors: | Wagner, T, Kretsinger, R.H, Bauerle, R, Tolbert, W.D. | | Deposit date: | 2000-08-04 | | Release date: | 2000-10-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 3-Deoxy-D-manno-octulosonate-8-phosphate synthase from Escherichia coli. Model of binding of phosphoenolpyruvate and D-arabinose-5-phosphate.
J.Mol.Biol., 301, 2000
|
|
1ALU
 
 | | HUMAN INTERLEUKIN-6 | | Descriptor: | INTERLEUKIN-6, L(+)-TARTARIC ACID, SULFATE ION | | Authors: | Somers, W.S, Stahl, M, Seehra, J.S. | | Deposit date: | 1997-06-03 | | Release date: | 1998-06-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 A crystal structure of interleukin 6: implications for a novel mode of receptor dimerization and signaling.
EMBO J., 16, 1997
|
|
8BAO
 
 | |
