1FK7
 
 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH RICINOLEIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | 分子名称: | FORMIC ACID, NON-SPECIFIC LIPID TRANSFER PROTEIN, RICINOLEIC ACID | | 著者 | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | 登録日 | 2000-08-09 | | 公開日 | 2001-06-06 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
6HSV
 
 | | Engineered higher-order assembly of Cholera Toxin B subunits via the addition of C-terminal parallel coiled-coiled domains | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, PHOSPHATE ION, ... | | 著者 | Pearson, A.R, Turnbull, W.B, Ross, J.F, Trinh, C.H, Webb, M.E. | | 登録日 | 2018-10-01 | | 公開日 | 2019-03-20 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Directed Assembly of Homopentameric Cholera Toxin B-Subunit Proteins into Higher-Order Structures Using Coiled-Coil Appendages.
J.Am.Chem.Soc., 141, 2019
|
|
1FKI
 
 | | DESIGN, SYNTHESIS, AND KINETIC EVALUATION OF HIGH-AFFINITY FKBP LIGANDS, AND THE X-RAY CRYSTAL STRUCTURES OF THEIR COMPLEXES WITH FKBP12 | | 分子名称: | (21S)-1AZA-4,4-DIMETHYL-6,19-DIOXA-2,3,7,20-TETRAOXOBICYCLO[19.4.0] PENTACOSANE, FK506 BINDING PROTEIN | | 著者 | Holt, D.A, Luengo, J.I, Yamashita, D.S, Oh, H.-J, Konialian, A.L, Yen, H.-K, Rozamus, L.W, Brandt, M, Bossard, M.J, Levy, M.A, Eggleston, D.S, Stout, T.J, Liang, J, Schultz, L.W, Clardy, J. | | 登録日 | 1993-08-05 | | 公開日 | 1994-01-31 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | DESIGN, SYNTHESIS, AND KINETIC EVALUATION OF HIGH-AFFINITY FKBP LIGANDS AND THE X-RAY CRYSTAL-STRUCTURES OF THEIR COMPLEXES WITH FKBP12.
J.Am.Chem.Soc., 115, 1993
|
|
4N39
 
 | |
8TNE
 
 | | Crystal structure of bacterial pectin methylesterase Pme8A from rumen Butyrivibrio | | 分子名称: | 1,2-ETHANEDIOL, Pectinesterase | | 著者 | Carbone, V, Reilly, K, Sang, C, Schofield, L, Ronimus, R, Kelly, W.J, Attwood, G.T, Palevich, N. | | 登録日 | 2023-08-01 | | 公開日 | 2023-08-16 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal Structures of Bacterial Pectin Methylesterases Pme8A and PmeC2 from Rumen Butyrivibrio .
Int J Mol Sci, 24, 2023
|
|
2V7Y
 
 | | Crystal structure of the molecular chaperone DnaK from Geobacillus kaustophilus HTA426 in post-ATP hydrolysis state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, CHAPERONE PROTEIN DNAK, MAGNESIUM ION, ... | | 著者 | Chang, Y.-W, Sun, Y.-J, Wang, C, Hsiao, C.-D. | | 登録日 | 2007-08-02 | | 公開日 | 2008-04-08 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.37 Å) | | 主引用文献 | Crystal Structures of the 70-kDa Heat Shock Proteins in Domain Disjoining Conformation.
J.Biol.Chem., 283, 2008
|
|
1FGT
 
 | |
7FC3
 
 | | structure of NL63 receptor-binding domain complexed with horse ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | 著者 | Wang, X.Q, Ge, J.W, Lan, J. | | 登録日 | 2021-07-13 | | 公開日 | 2021-09-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.19 Å) | | 主引用文献 | Structural insights into the binding of SARS-CoV-2, SARS-CoV, and hCoV-NL63 spike receptor-binding domain to horse ACE2.
Structure, 30, 2022
|
|
7ZUB
 
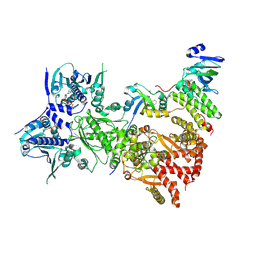 | | Cryo-EM structure of the indirubin-bound Hsp90-XAP2-AHR complex | | 分子名称: | (3~{Z})-3-(3-oxidanylidene-1~{H}-indol-2-ylidene)-1~{H}-indol-2-one, ADENOSINE-5'-DIPHOSPHATE, AH receptor-interacting protein, ... | | 著者 | Gruszczyk, J, Savva, C.G, Lai-Kee-Him, J, Bous, J, Ancelin, A, Kwong, H.S, Grandvuillemin, L, Bourguet, W. | | 登録日 | 2022-05-12 | | 公開日 | 2022-11-23 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.85 Å) | | 主引用文献 | Cryo-EM structure of the agonist-bound Hsp90-XAP2-AHR cytosolic complex.
Nat Commun, 13, 2022
|
|
4N9B
 
 | | Fragment-based Design of 3-Aminopyridine-derived Amides as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | 分子名称: | 1-methyl-N-(pyridin-3-yl)-1H-pyrazole-5-carboxamide, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | 著者 | Dragovich, P.S, Zhao, G, Baumeister, T, Bravo, B, Giannetti, A.M, Ho, Y, Hua, R, Li, G, Liang, X, O'Brien, T, Skelton, N.J, Wang, C, Zhai, Q, Oh, A, Wang, W, Wang, Y, Xiao, Y, Yuen, P, Zak, M, Zheng, X. | | 登録日 | 2013-10-20 | | 公開日 | 2014-02-19 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.859 Å) | | 主引用文献 | Fragment-based design of 3-aminopyridine-derived amides as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 24, 2014
|
|
7EW0
 
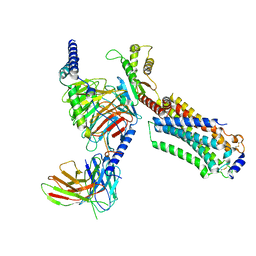 | | Cryo-EM structure of ozanimod -bound Sphingosine-1-phosphate receptor 1 in complex with Gi protein | | 分子名称: | 5-[3-[(1~{S})-1-(2-hydroxyethylamino)-2,3-dihydro-1~{H}-inden-4-yl]-1,2,4-oxadiazol-5-yl]-2-propan-2-yloxy-benzenecarbonitrile, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Yuan, Y, Jia, G.W, Su, Z.M, Shao, Z.H. | | 登録日 | 2021-05-24 | | 公開日 | 2021-09-29 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (3.42 Å) | | 主引用文献 | Structures of signaling complexes of lipid receptors S1PR1 and S1PR5 reveal mechanisms of activation and drug recognition.
Cell Res., 31, 2021
|
|
8A55
 
 | |
7EW7
 
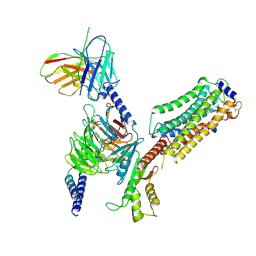 | | Cryo-EM structure of SEW2871-bound Sphingosine-1-phosphate receptor 1 in complex with Gi protein | | 分子名称: | 5-[4-phenyl-5-(trifluoromethyl)thiophen-2-yl]-3-[3-(trifluoromethyl)phenyl]-1,2,4-oxadiazole, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Jia, G.W, Yuan, Y, Su, Z.M, Shao, Z.H. | | 登録日 | 2021-05-24 | | 公開日 | 2021-09-29 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (3.27 Å) | | 主引用文献 | Structures of signaling complexes of lipid receptors S1PR1 and S1PR5 reveal mechanisms of activation and drug recognition.
Cell Res., 31, 2021
|
|
1F8N
 
 | |
6DOG
 
 | |
6DPC
 
 | |
3NIP
 
 | | Crystal structure of Pseudomonas aeruginosa guanidinopropionase complexed with 1,6-diaminohexane | | 分子名称: | 3-guanidinopropionase, HEXANE-1,6-DIAMINE | | 著者 | Lee, S.J, Kim, H.S, Kim, D.J, Yoon, H.J, Kim, K.H, Yoon, J.Y, Jang, J.Y, Im, H, An, D, Suh, S.W. | | 登録日 | 2010-06-16 | | 公開日 | 2011-06-01 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structures of Pseudomonas aeruginosa guanidinobutyrase and guanidinopropionase, members of the ureohydrolase superfamily
J.Struct.Biol., 175, 2011
|
|
6DOU
 
 | |
7EW2
 
 | | Cryo-EM structure of pFTY720-bound Sphingosine 1-phosphate receptor 3 in complex with Gi protein | | 分子名称: | (2~{S})-2-azanyl-4-(4-octylphenyl)-2-[[oxidanyl-bis(oxidanylidene)-$l^{6}-phosphanyl]oxymethyl]butan-1-ol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Zhao, C, Wang, W, Wang, H.L, Shao, Z.H. | | 登録日 | 2021-05-24 | | 公開日 | 2021-09-29 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural insights into sphingosine-1-phosphate recognition and ligand selectivity of S1PR3-Gi signaling complexes.
Cell Res., 32, 2022
|
|
6DPN
 
 | |
1FD7
 
 | | HEAT-LABILE ENTEROTOXIN B-PENTAMER WITH BOUND LIGAND BMSC001 | | 分子名称: | HEAT-LABILE ENTEROTOXIN B CHAIN, N-BENZYL-3-(ALPHA-D-GALACTOS-1-YL)-BENZAMIDE | | 著者 | Fan, E, Merritt, E.A, Pickens, J, Ahn, M, Hol, W.G.J. | | 登録日 | 2000-07-19 | | 公開日 | 2000-08-10 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Exploration of the GM1 receptor-binding site of heat-labile enterotoxin and cholera toxin by phenyl-ring-containing galactose derivatives.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
2VR8
 
 | | Crystal Structure of G85R ALS mutant of Human Cu,Zn Superoxide Dismutase (CuZnSOD) at 1.36 A resolution | | 分子名称: | COPPER (II) ION, SULFATE ION, SUPEROXIDE DISMUTASE [CU-ZN], ... | | 著者 | Antonyuk, S, Cao, X, Seetharaman, S.V, Whitson, L.J, Taylor, A.B, Holloway, S.P, Strange, R.W, Doucette, P.A, Tiwari, A, Hayward, L.J, Padua, S, Cohlberg, J.A, Selverstone Valentine, J, Hasnain, S.S, Hart, P.J. | | 登録日 | 2008-03-28 | | 公開日 | 2008-04-08 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | Structures of the G85R Variant of Sod1 in Familial Amyotrophic Lateral Sclerosis.
J.Biol.Chem., 283, 2008
|
|
1FEH
 
 | | FE-ONLY HYDROGENASE FROM CLOSTRIDIUM PASTEURIANUM | | 分子名称: | 2 IRON/2 SULFUR/5 CARBONYL/2 WATER INORGANIC CLUSTER, FE2/S2 (INORGANIC) CLUSTER, IRON/SULFUR CLUSTER, ... | | 著者 | Peters, J.W, Lanzilotta, W.N, Lemon, B.J, Seefeldt, L.C. | | 登録日 | 1998-10-28 | | 公開日 | 1999-01-13 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | X-ray crystal structure of the Fe-only hydrogenase (CpI) from Clostridium pasteurianum to 1.8 angstrom resolution.
Science, 282, 1998
|
|
3PIM
 
 | | Crystal structure of Mxr1 from Saccharomyces cerevisiae in unusual oxidized form | | 分子名称: | Peptide methionine sulfoxide reductase | | 著者 | Ma, X.X, Guo, P.C, Shi, W.W, Luo, M, Tan, X.F, Chen, Y, Zhou, C.Z. | | 登録日 | 2010-11-07 | | 公開日 | 2011-02-23 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural plasticity of the thioredoxin recognition site of yeast methionine S-sulfoxide reductase Mxr1
J.Biol.Chem., 286, 2011
|
|
7EW1
 
 | | Cryo-EM structure of siponimod -bound Sphingosine-1-phosphate receptor 5 in complex with Gi protein | | 分子名称: | 1-[[4-[(~{E})-~{N}-[[4-cyclohexyl-3-(trifluoromethyl)phenyl]methoxy]-~{C}-methyl-carbonimidoyl]-2-ethyl-phenyl]methyl]azetidine-3-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Yuan, Y, Jia, G.W, Shao, Z.H, Su, Z.M. | | 登録日 | 2021-05-24 | | 公開日 | 2021-09-29 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structures of signaling complexes of lipid receptors S1PR1 and S1PR5 reveal mechanisms of activation and drug recognition.
Cell Res., 31, 2021
|
|
