1Q44
 
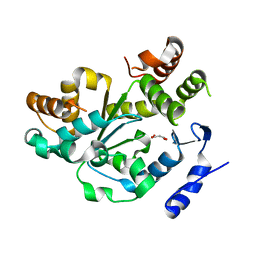 | | Crystal Structure of an Arabidopsis Thaliana Putative Steroid Sulfotransferase | | 分子名称: | MALONIC ACID, Steroid Sulfotransferase | | 著者 | Phillips Jr, G.N, Smith, D.W, Johnson, K.A, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | 登録日 | 2003-08-01 | | 公開日 | 2003-11-25 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of At2g03760, a putative steroid sulfotransferase from Arabidopsis thaliana
Proteins, 57, 2004
|
|
2X3T
 
 | | Glutaraldehyde-crosslinked wheat germ agglutinin isolectin 1 crystal soaked with a synthetic glycopeptide | | 分子名称: | 2-acetamido-1-O-carbamoyl-2-deoxy-alpha-D-glucopyranose, AGGLUTININ ISOLECTIN 1, D-ALPHA-AMINOBUTYRIC ACID, ... | | 著者 | Schwefel, D, Maierhofer, C, Wittmann, V, Diederichs, K, Welte, W. | | 登録日 | 2010-01-26 | | 公開日 | 2010-06-30 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.749 Å) | | 主引用文献 | Structural Basis of Multivalent Binding to Wheat Germ Agglutinin.
J.Am.Chem.Soc., 132, 2010
|
|
4MW1
 
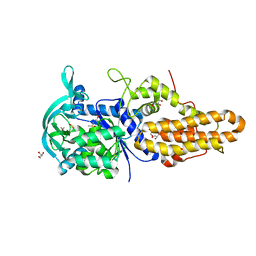 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor 1-{3-[(3-chloro-5-methoxybenzyl)amino]propyl}-3-thiophen-3-ylurea (Chem 1444) | | 分子名称: | 1-{3-[(3-chloro-5-methoxybenzyl)amino]propyl}-3-thiophen-3-ylurea, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Koh, C.Y, Kim, J.E, Wetzel, A.B, de van der Schueren, W.J, Shibata, S, Liu, J, Zhang, Z, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | 登録日 | 2013-09-24 | | 公開日 | 2014-04-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.494 Å) | | 主引用文献 | Structures of Trypanosoma brucei Methionyl-tRNA Synthetase with Urea-Based Inhibitors Provide Guidance for Drug Design against Sleeping Sickness.
Plos Negl Trop Dis, 8, 2014
|
|
3DL6
 
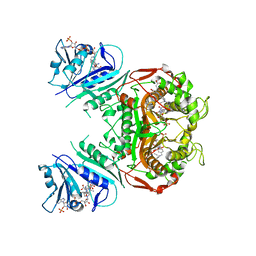 | | Crystal Structure of the A287F/S290G Active Site Mutant of TS-DHFR from Cryptosporidium hominis | | 分子名称: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, DIHYDROFOLIC ACID, ... | | 著者 | Martucci, W.E, Vargo, M.A, Anderson, K.S. | | 登録日 | 2008-06-26 | | 公開日 | 2008-08-19 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (3.25 Å) | | 主引用文献 | Explaining an unusually fast parasitic enzyme: folate tail-binding residues dictate substrate positioning and catalysis in Cryptosporidium hominis thymidylate synthase.
Biochemistry, 47, 2008
|
|
2X1C
 
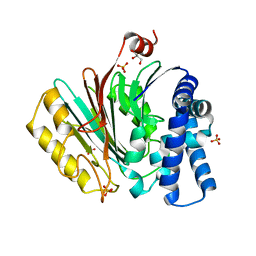 | | The crystal structure of precursor acyl coenzyme A:isopenicillin N acyltransferase from Penicillium chrysogenum | | 分子名称: | ACYL-COENZYME, CHLORIDE ION, GLYCEROL, ... | | 著者 | Bokhove, M, Yoshida, H, Hensgens, C.M.H, van der Laan, J.M, Sutherland, J.D, Dijkstra, B.W. | | 登録日 | 2009-12-23 | | 公開日 | 2010-03-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structures of an Isopenicillin N Converting Ntn-Hydrolase Reveal Different Catalytic Roles for the Active Site Residues of Precursor and Mature Enzyme.
Structure, 18, 2010
|
|
1D97
 
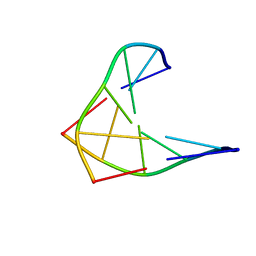 | | CHIRAL PHOSPHOROTHIOATE ANALOGUES OF B-DNA: THE CRYSTAL STRUCTURE OF RP-D(GP(S) CPGP(S)CPGP(S)C) | | 分子名称: | DNA (5'-D(RP*GP*(SC)P*GP*(SC)P*GP*(SC))-3') | | 著者 | Cruse, W.B.T, Salisbury, S.A, Brown, T, Cosstick, R, Eckstein, F, Kennard, O. | | 登録日 | 1992-10-17 | | 公開日 | 1993-07-15 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Chiral phosphorothioate analogues of B-DNA. The crystal structure of Rp-d[Gp(S)CpGp(S)CpGp(S)C].
J.Mol.Biol., 192, 1986
|
|
1U8R
 
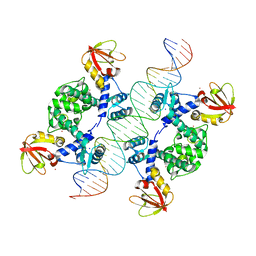 | | Crystal Structure of an IdeR-DNA Complex Reveals a Conformational Change in Activated IdeR for Base-specific Interactions | | 分子名称: | COBALT (II) ION, Iron-dependent repressor ideR, SODIUM ION, ... | | 著者 | Wisedchaisri, G, Holmes, R.K, Hol, W.G.J. | | 登録日 | 2004-08-06 | | 公開日 | 2004-10-05 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Crystal Structure of an IdeR-DNA Complex Reveals a Conformational Change in Activated IdeR for Base-specific Interactions.
J.Mol.Biol., 342, 2004
|
|
6D0P
 
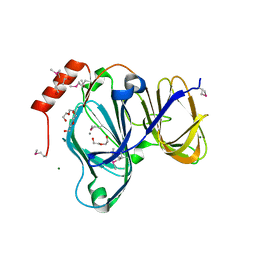 | | 1.88 Angstrom Resolution Crystal Structure of Quercetin 2,3-dioxygenase from Acinetobacter baumannii | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Minasov, G, Shuvalova, L, Brunzelle, J.S, Dubrovska, I, Kiryukhina, O, Endres, M, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2018-04-10 | | 公開日 | 2018-04-25 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | 1.88 Angstrom Resolution Crystal Structure of Quercetin 2,3-dioxygenase from Acinetobacter baumannii
To be Published
|
|
1RQC
 
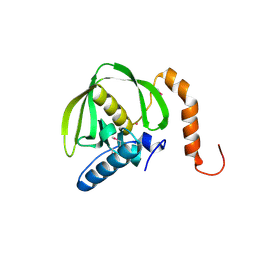 | | Crystals of peptide deformylase from Plasmodium falciparum with ten subunits per asymmetric unit reveal critical characteristics of the active site for drug design | | 分子名称: | COBALT (II) ION, formylmethionine deformylase | | 著者 | Robien, M.A, Nguyen, K.T, Kumar, A, Hirsh, I, Turley, S, Pei, D, Hol, W.G. | | 登録日 | 2003-12-04 | | 公開日 | 2004-01-20 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | An improved crystal form of Plasmodium falciparum peptide deformylase
Protein Sci., 13, 2004
|
|
1RHU
 
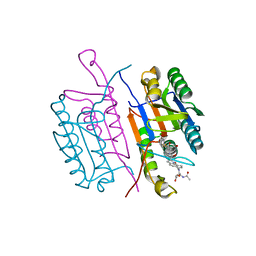 | | CRYSTAL STRUCTURE OF THE COMPLEX OF CASPASE-3 WITH A 5,6,7 TRICYCLIC PEPTIDOMIMETIC INHIBITOR | | 分子名称: | (3S)-3-[({(2S)-5-[(N-ACETYL-L-ALPHA-ASPARTYL)AMINO]-4-OXO-1,2,4,5,6,7-HEXAHYDROAZEPINO[3,2,1-HI]INDOL-2-YL}CARBONYL)AMINO]-5-(BENZYLSULFANYL)-4-OXOPENTANOIC ACID, Caspase-3 | | 著者 | Becker, J.W, Rotonda, J, Soisson, S.M. | | 登録日 | 2003-11-14 | | 公開日 | 2004-05-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | Reducing the Peptidyl Features of Caspase-3 Inhibitors: A Structural Analysis.
J.Med.Chem., 47, 2004
|
|
6I53
 
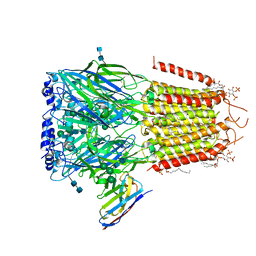 | | Cryo-EM structure of the human synaptic alpha1-beta3-gamma2 GABAA receptor in complex with Megabody38 in a lipid nanodisc | | 分子名称: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit alpha-1, ... | | 著者 | Laverty, D, Desai, R, Uchanski, T, Masiulis, S, Wojciech, J.S, Malinauskas, T, Zivanov, J, Pardon, E, Steyaert, J, Miller, K.W, Aricescu, A.R. | | 登録日 | 2018-11-12 | | 公開日 | 2019-01-02 | | 最終更新日 | 2022-03-30 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM structure of the human alpha 1 beta 3 gamma 2 GABAAreceptor in a lipid bilayer.
Nature, 565, 2019
|
|
5DOY
 
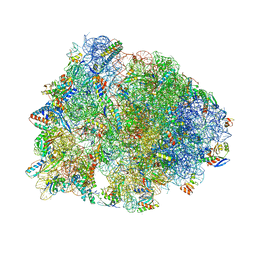 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with antibiotic Hygromycin A, mRNA and three tRNAs in the A, P and E sites at 2.6A resolution | | 分子名称: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | 著者 | Polikanov, Y.S, Starosta, A.L, Juette, M.F, Altman, R.B, Terry, D.S, Lu, W, Burnett, B.J, Dinos, G, Reynolds, K, Blanchard, S.C, Steitz, T.A, Wilson, D.N. | | 登録日 | 2015-09-11 | | 公開日 | 2015-12-30 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Distinct tRNA Accommodation Intermediates Observed on the Ribosome with the Antibiotics Hygromycin A and A201A.
Mol.Cell, 58, 2015
|
|
1CSB
 
 | |
3HJV
 
 | | 1.7 Angstrom resolution crystal structure of an acyl carrier protein S-malonyltransferase from Vibrio cholerae O1 biovar eltor str. N16961 | | 分子名称: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, CHLORIDE ION, Malonyl Coa-acyl carrier protein transacylase, ... | | 著者 | Halavaty, A.S, Wawrzak, Z, Anderson, S, Skarina, T, Onopriyenko, O, Kwon, K, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2009-05-22 | | 公開日 | 2009-06-09 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | 1.7 Angstrom resolution crystal structure of an acyl carrier protein S-malonyltransferase from Vibrio cholerae O1 biovar eltor str. N16961
To be Published
|
|
1DYE
 
 | |
1PPR
 
 | | PERIDININ-CHLOROPHYLL-PROTEIN OF AMPHIDINIUM CARTERAE | | 分子名称: | CHLOROPHYLL A, DIGALACTOSYL DIACYL GLYCEROL (DGDG), PERIDININ, ... | | 著者 | Hofmann, E, Welte, W, Diederichs, K. | | 登録日 | 1996-03-06 | | 公開日 | 1997-08-20 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis of light harvesting by carotenoids: peridinin-chlorophyll-protein from Amphidinium carterae.
Science, 272, 1996
|
|
1PMA
 
 | | PROTEASOME FROM THERMOPLASMA ACIDOPHILUM | | 分子名称: | PROTEASOME | | 著者 | Loewe, J, Stock, D, Jap, B, Zwickl, P, Baumeister, W, Huber, R. | | 登録日 | 1994-12-19 | | 公開日 | 1996-06-20 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution.
Science, 268, 1995
|
|
2WHE
 
 | | Structure of native Beta-Phosphoglucomutase in an open conformation without bound ligands. | | 分子名称: | BETA-PHOSPHOGLUCOMUTASE, MAGNESIUM ION | | 著者 | Bowler, M.W, Baxter, N.J, Webster, C.E, Pollard, S, Alizadeh, T, Hounslow, A.M, Cliff, M.J, Bermel, W, Williams, N.H, Hollfelder, F, Blackburn, G.M, Waltho, J.P. | | 登録日 | 2009-05-04 | | 公開日 | 2009-09-15 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Atomic Details of Near-Transition State Conformers for Enzyme Phosphoryl Transfer Revealed by Mgf-3 Rather Than by Phosphoranes.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1PWY
 
 | | CRYSTAL STRUCTURE OF HUMAN PNP COMPLEXED WITH ACYCLOVIR | | 分子名称: | 9-HYROXYETHOXYMETHYLGUANINE, Purine nucleoside phosphorylase, SULFATE ION | | 著者 | Dos Santos, D.M, Canduri, F, Pereira, J.H, Vinicius Bertacine Dias, M, Silva, R.G, Mendes, M.A, Palma, M.S, Basso, L.A, De Azevedo, W.F, Santos, D.S. | | 登録日 | 2003-07-02 | | 公開日 | 2004-03-23 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of human purine nucleoside phosphorylase complexed with acyclovir.
Biochem.Biophys.Res.Commun., 308, 2003
|
|
1DYB
 
 | |
4N9E
 
 | | Fragment-based Design of 3-Aminopyridine-derived Amides as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | 分子名称: | 1,2-ETHANEDIOL, 1-[(1-benzoylpiperidin-4-yl)methyl]-N-(pyridin-3-yl)-1H-benzimidazole-5-carboxamide, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ... | | 著者 | Dragovich, P.S, Zhao, G, Baumeister, T, Bravo, B, Giannetti, A.M, Ho, Y, Hua, R, Li, G, Liang, X, O'Brien, T, Skelton, N.J, Wang, C, Zhao, Q, Oh, A, Wang, W, Wang, Y, Xiao, Y, Yuen, P, Zak, M, Zheng, X. | | 登録日 | 2013-10-20 | | 公開日 | 2014-02-19 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Fragment-based design of 3-aminopyridine-derived amides as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 24, 2014
|
|
6B0D
 
 | | An E. coli DPS protein from ferritin superfamily | | 分子名称: | DNA protection during starvation protein, FORMIC ACID, SODIUM ION | | 著者 | Rui, W, Ruslan, S, Ronan, K, Adam, J.S. | | 登録日 | 2017-09-14 | | 公開日 | 2018-09-19 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | SIMBAD: a sequence-independent molecular-replacement pipeline.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
4DLM
 
 | | Crystal structure of an amidohydrolase (COG3618) from burkholderia multivorans (TARGET EFI-500235) with bound ZN, space group P212121 | | 分子名称: | Amidohydrolase 2, ZINC ION | | 著者 | Vetting, M.W, Toro, R, Bhosle, R, Seidel, R.D, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Al Obaidi, N.F, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Raushel, F.M, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2012-02-06 | | 公開日 | 2012-02-29 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.925 Å) | | 主引用文献 | Crystal structure of an amidohydrolase (COG3618) from burkholderia multivorans (TARGET EFI-500235) with bound ZN, space group P212121
to be published
|
|
3GED
 
 | | Fingerprint and Structural Analysis of a Apo SCOR enzyme from Clostridium thermocellum | | 分子名称: | GLYCEROL, SODIUM ION, Short-chain dehydrogenase/reductase SDR, ... | | 著者 | Huether, R, Liu, Z.J, Xu, H, Wang, B.C, Pletnev, V, Mao, Q, Umland, T, Duax, W. | | 登録日 | 2009-02-25 | | 公開日 | 2009-03-17 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.698 Å) | | 主引用文献 | Sequence fingerprint and structural analysis of the SCOR enzyme A3DFK9 from Clostridium thermocellum.
Proteins, 78, 2010
|
|
3GEU
 
 | | Crystal Structure of IcaR from Staphylococcus aureus, a member of the tetracycline repressor protein family | | 分子名称: | CHLORIDE ION, FORMIC ACID, Intercellular adhesion protein R, ... | | 著者 | Anderson, S.M, Brunzelle, J.S, Wawrzak, Z, Skarina, T, Papazisi, L, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2009-02-26 | | 公開日 | 2009-03-10 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal Structure of IcaR from Staphylococcus aureus, a member of the tetracycline repressor protein family
To be Published
|
|
