4MHE
 
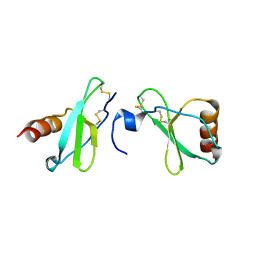 | | Crystal structure of CC-chemokine 18 | | 分子名称: | ACETATE ION, C-C motif chemokine 18 | | 著者 | Liang, W.G, Tang, W.-J. | | 登録日 | 2013-08-29 | | 公開日 | 2014-09-03 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structures of human CCL18, CCL3, and CCL4 reveal molecular determinants for quaternary structures and sensitivity to insulin-degrading enzyme.
J.Mol.Biol., 427, 2015
|
|
8IHS
 
 | | Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 in complex with ochratoxin A | | 分子名称: | (2~{S})-2-[[(3~{R})-5-chloranyl-3-methyl-8-oxidanyl-1-oxidanylidene-3,4-dihydroisochromen-7-yl]carbonylamino]-3-phenyl-propanoic acid, Amidohydrolase family protein, ZINC ION | | 著者 | Dai, L.H, Niu, D, Huang, J.-W, Li, X, Shen, P.P, Li, H, Hu, Y.M, Yang, Y, Chen, C.-C, Guo, R.-T. | | 登録日 | 2023-02-23 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase.
J Hazard Mater, 458, 2023
|
|
4QUY
 
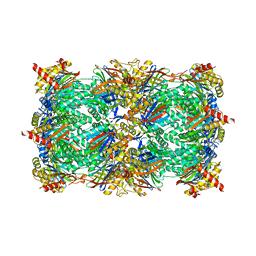 | | yCP beta5-A49S-mutant | | 分子名称: | CHLORIDE ION, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | 著者 | Huber, E.M, Heinemeyer, W, Groll, M. | | 登録日 | 2014-07-14 | | 公開日 | 2015-02-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
3S73
 
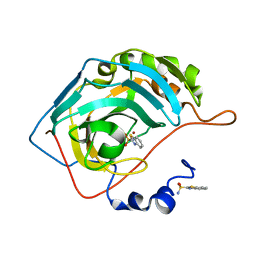 | | The origin of the hydrophobic effect in the molecular recognition of arylsulfonamides by carbonic anhydrase | | 分子名称: | 1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | 著者 | Snyder, P.W, Heroux, A, Whitesides, G.W. | | 登録日 | 2011-05-26 | | 公開日 | 2011-12-21 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Mechanism of the hydrophobic effect in the biomolecular recognition of arylsulfonamides by carbonic anhydrase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4QW3
 
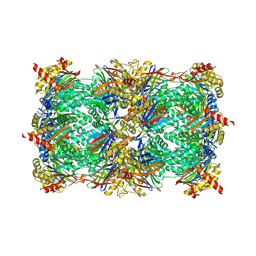 | | yCP beta5-C63F mutant in complex with bortezomib | | 分子名称: | CHLORIDE ION, MAGNESIUM ION, N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, ... | | 著者 | Huber, E.M, Heinemeyer, W, Groll, M. | | 登録日 | 2014-07-16 | | 公開日 | 2015-02-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
5QPS
 
 | | PanDDA analysis group deposition -- Crystal Structure of T. cruzi FPPS in complex with FMOPL000644a | | 分子名称: | ACETATE ION, Farnesyl diphosphate synthase, SULFATE ION, ... | | 著者 | Petrick, J.K, Nelson, E.R, Muenzker, L, Krojer, T, Douangamath, A, Brandao-Neto, J, von Delft, F, Dekker, C, Jahnke, W. | | 登録日 | 2019-03-12 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.61 Å) | | 主引用文献 | PanDDA analysis group deposition - FPPS screened against the DSI Fragment Library
To Be Published
|
|
5ZAP
 
 | | Atomic structure of the herpes simplex virus type 2 B-capsid | | 分子名称: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | 著者 | Yuan, S, Wang, J.L, Zhu, D.J, Wang, N, Gao, Q, Chen, W.Y, Tang, H, Wang, J.Z, Zhang, X.Z, Liu, H.R, Rao, Z.H, Wang, X.X. | | 登録日 | 2018-02-08 | | 公開日 | 2018-04-18 | | 最終更新日 | 2018-09-05 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Cryo-EM structure of a herpesvirus capsid at 3.1 angstrom.
Science, 360, 2018
|
|
8IHR
 
 | | Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 in complex with Phe | | 分子名称: | Amidohydrolase family protein, PHENYLALANINE, ZINC ION | | 著者 | Dai, L.H, Niu, D, Huang, J.-W, Li, X, Shen, P.P, Li, H, Hu, Y.M, Yang, Y, Chen, C.-C, Guo, R.-T. | | 登録日 | 2023-02-23 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase.
J Hazard Mater, 458, 2023
|
|
5QQ3
 
 | | PanDDA analysis group deposition -- Crystal Structure of T. cruzi FPPS in complex with FMOPL000672a | | 分子名称: | ACETATE ION, Farnesyl diphosphate synthase, SULFATE ION, ... | | 著者 | Petrick, J.K, Nelson, E.R, Muenzker, L, Krojer, T, Douangamath, A, Brandao-Neto, J, von Delft, F, Dekker, C, Jahnke, W. | | 登録日 | 2019-03-12 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | PanDDA analysis group deposition - FPPS screened against the DSI Fragment Library
To Be Published
|
|
8EC0
 
 | | III2IV respiratory supercomplex from Saccharomyces cerevisiae cardiolipin-lacking mutant | | 分子名称: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, 5-(3,7,11,15,19,23-HEXAMETHYL-TETRACOSA-2,6,10,14,18,22-HEXAENYL)-2,3-DIMETHOXY-6-METHYL-BENZENE-1,4-DIOL, ... | | 著者 | Hryc, C.F, Mileykovskaya, E, Baker, M, Dowhan, W. | | 登録日 | 2022-08-31 | | 公開日 | 2023-05-24 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural insights into cardiolipin replacement by phosphatidylglycerol in a cardiolipin-lacking yeast respiratory supercomplex.
Nat Commun, 14, 2023
|
|
5QQA
 
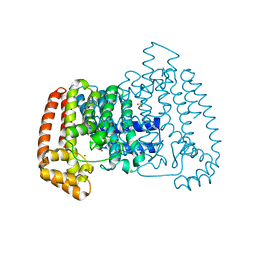 | | PanDDA analysis group deposition -- Crystal Structure of T. cruzi FPPS in complex with FMOOA000648a | | 分子名称: | ACETATE ION, Farnesyl diphosphate synthase, SULFATE ION, ... | | 著者 | Petrick, J.K, Nelson, E.R, Muenzker, L, Krojer, T, Douangamath, A, Brandao-Neto, J, von Delft, F, Dekker, C, Jahnke, W. | | 登録日 | 2019-03-12 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | PanDDA analysis group deposition - FPPS screened against the DSI Fragment Library
To Be Published
|
|
5QPK
 
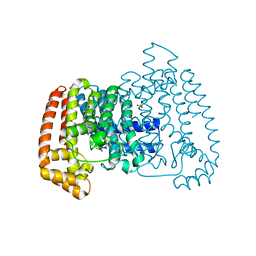 | | PanDDA analysis group deposition -- Crystal Structure of T. cruzi FPPS in complex with FMOPL000586a | | 分子名称: | ACETATE ION, Farnesyl diphosphate synthase, SULFATE ION, ... | | 著者 | Petrick, J.K, Nelson, E.R, Muenzker, L, Krojer, T, Douangamath, A, Brandao-Neto, J, von Delft, F, Dekker, C, Jahnke, W. | | 登録日 | 2019-03-12 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | PanDDA analysis group deposition - FPPS screened against the DSI Fragment Library
To Be Published
|
|
8IHQ
 
 | | Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 | | 分子名称: | Amidohydrolase family protein, ZINC ION | | 著者 | Dai, L.H, Niu, D, Huang, J.-W, Li, X, Shen, P.P, Li, H, Hu, Y.M, Yang, Y, Chen, C.-C, Guo, R.-T. | | 登録日 | 2023-02-23 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (2.71 Å) | | 主引用文献 | Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase.
J Hazard Mater, 458, 2023
|
|
4HGU
 
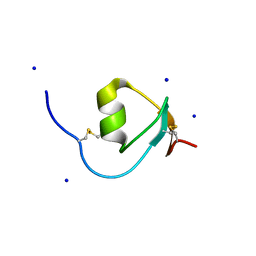 | | Crystal Structure of Galleria mellonella Silk Protease Inhibitor 2 | | 分子名称: | SODIUM ION, Silk protease inhibitor 2 | | 著者 | Krzywda, S, Jaskolski, M, Dvornyk, A, Kludkiewicz, B, Grzelak, K, Zagorski, W, Bal, W, Kopera, E. | | 登録日 | 2012-10-08 | | 公開日 | 2013-10-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (0.98 Å) | | 主引用文献 | Atomic resolution structure of a protein prepared by non-enzymatic His-tag removal. Crystallographic and NMR study of GmSPI-2 inhibitor.
Plos One, 9, 2014
|
|
5QQC
 
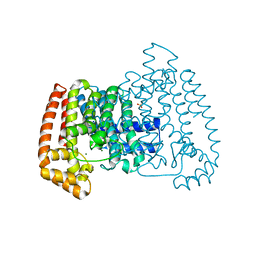 | | Crystal Structure of T. cruzi FPPS after initial refinement with no ligand modelled (structure $n) | | 分子名称: | ACETATE ION, Farnesyl diphosphate synthase, MAGNESIUM ION, ... | | 著者 | Petrick, J.K, Nelson, E.R, Muenzker, L, Krojer, T, Douangamath, A, Brandao-Neto, J, von Delft, F, Dekker, C, Jahnke, W. | | 登録日 | 2019-03-14 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | PanDDA analysis group deposition of ground-state model - FPPS screened against the DSI Fragment Library
To Be Published
|
|
4QZ3
 
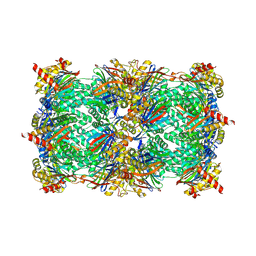 | | yCP beta5-A49V mutant in complex with the epoxyketone inhibitor ONX 0914 | | 分子名称: | 1,2,4-trideoxy-4-methyl-2-{[N-(morpholin-4-ylacetyl)-L-alanyl-O-methyl-L-tyrosyl]amino}-1-phenyl-D-xylitol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | 著者 | Huber, E.M, Heinemeyer, W, Groll, M. | | 登録日 | 2014-07-27 | | 公開日 | 2015-02-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
8ILA
 
 | | Crystal structure of LmbT from Streptomyces lincolnensis NRRL ISP-5355 in complex with substrates | | 分子名称: | (2~{S})-3-[2-[(2~{S},3~{R},4~{S},5~{R},6~{R})-6-[(1~{R},2~{R})-1-azanyl-2-oxidanyl-propyl]-3,4,5-tris(oxidanyl)oxan-2-yl]sulfanyl-1~{H}-imidazol-5-yl]-2-(trimethyl-$l^{4}-azanyl)propanoic acid, GUANOSINE-5'-DIPHOSPHATE, Glycosyltransferase | | 著者 | Dai, Y, Qiao, H, Xia, M, Fang, P, Liu, W. | | 登録日 | 2023-03-03 | | 公開日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Structural Basis of Low-Molecular-Weight Thiol Glycosylation in Lincomycin A Biosynthesis.
Acs Chem.Biol., 18, 2023
|
|
4QZ4
 
 | | yCP beta5-A49S mutant in complex with the epoxyketone inhibitor ONX 0914 | | 分子名称: | 1,2,4-trideoxy-4-methyl-2-{[N-(morpholin-4-ylacetyl)-L-alanyl-O-methyl-L-tyrosyl]amino}-1-phenyl-D-xylitol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | 著者 | Huber, E.M, Heinemeyer, W, Groll, M. | | 登録日 | 2014-07-27 | | 公開日 | 2015-02-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
5YZE
 
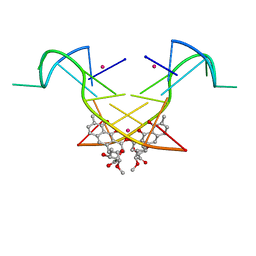 | | Crystal structure of the [Co2+-(chromomycin A3)2]-d(CCG)3 complex | | 分子名称: | (1S)-5-deoxy-1-O-methyl-1-C-[(2R,3S)-3,5,7,10-tetrahydroxy-6-methyl-4-oxo-1,2,3,4-tetrahydroanthracen-2-yl]-D-xylulose, 2,6-dideoxy-4-O-methyl-alpha-D-galactopyranose-(1-3)-(2R,3R,6R)-6-hydroxy-2-methyltetrahydro-2H-pyran-3-yl acetate, 3-C-methyl-4-O-acetyl-alpha-L-Olivopyranose-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol, ... | | 著者 | Hou, M.H, Chen, Y.W, Wu, P.C, Satange, R.B. | | 登録日 | 2017-12-14 | | 公開日 | 2018-10-17 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | CoII(Chromomycin)2 Complex Induces a Conformational Change of CCG Repeats from i-Motif to Base-Extruded DNA Duplex
Int J Mol Sci, 19, 2018
|
|
4I9N
 
 | | Crystal structure of rabbit LDHA in complex with AP28161 and AP28122 | | 分子名称: | 6-({2-[(5-chloro-4-{[(2S)-2,3-dihydroxypropyl]oxy}-2-methoxyphenyl)amino]-2-oxoethyl}sulfanyl)pyridine-3-carboxylic acid, 6-[3-(carboxymethoxy)-5-fluorophenyl]pyridine-3-carboxylic acid, L-lactate dehydrogenase A chain | | 著者 | Zhou, T, Kohlmann, A, Stephan, Z.G, Li, F, Commodore, L, Greenfield, M.T, Shakespeare, W.C, Zhu, X, Dalgarno, D.C. | | 登録日 | 2012-12-05 | | 公開日 | 2013-01-23 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Fragment growing and linking lead to novel nanomolar lactate dehydrogenase inhibitors.
J.Med.Chem., 56, 2013
|
|
4E5O
 
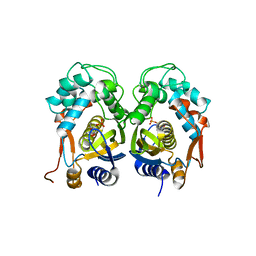 | | Crystal structure of mouse thymidylate synthase in complex with dUMP | | 分子名称: | 1,4-BUTANEDIOL, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Thymidylate synthase | | 著者 | Dowiercial, A, Jarmula, A, Rypniewski, W, Sokolowska, M, Fraczyk, T, Ciesla, J, Rode, W. | | 登録日 | 2012-03-14 | | 公開日 | 2012-04-18 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Mouse thymidylate synthase does not show the inactive conformation, observed for the human enzyme
Struct Chem, 2016
|
|
6EPD
 
 | | Substrate processing state 26S proteasome (SPS1) | | 分子名称: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 13, ... | | 著者 | Guo, Q, Lehmer, C, Martinez-Sanchez, A, Rudack, T, Beck, F, Hartmann, H, Hipp, M.S, Hartl, F.U, Edbauer, D, Baumeister, W, Fernandez-Busnadiego, R. | | 登録日 | 2017-10-11 | | 公開日 | 2018-02-07 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (15.4 Å) | | 主引用文献 | In Situ Structure of Neuronal C9orf72 Poly-GA Aggregates Reveals Proteasome Recruitment.
Cell, 172, 2018
|
|
4M7Y
 
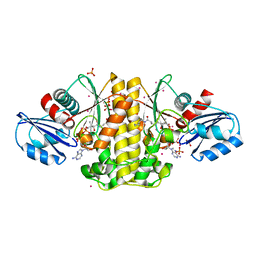 | | Staphylococcus aureus Type II pantothenate kinase in complex with a pantothenate analog | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-N-pentyl-beta-alaninamide, PHOSPHATE ION, ... | | 著者 | Mottaghi, K, Hong, B, Tempel, W, Park, H, Structural Genomics Consortium (SGC) | | 登録日 | 2013-08-12 | | 公開日 | 2014-09-03 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Discovery of Potent Pantothenamide Inhibitors of Staphylococcus aureus Pantothenate Kinase through a Minimal SAR Study: Inhibition Is Due to Trapping of the Product.
ACS Infect Dis, 2, 2016
|
|
5K9P
 
 | | Ser20 phosphorylated ubiquitin | | 分子名称: | Polyubiquitin-B | | 著者 | Huguenin-Dezot, N, Chin, J.W. | | 登録日 | 2016-06-01 | | 公開日 | 2016-07-20 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Synthesis of Isomeric Phosphoubiquitin Chains Reveals that Phosphorylation Controls Deubiquitinase Activity and Specificity.
Cell Rep, 16, 2016
|
|
4I6W
 
 | | 3-hydroxy-3-methylglutaryl (HMG) Coenzyme-A reductase complexed with thiomevalonate | | 分子名称: | (3S)-3-hydroxy-3-methyl-5-sulfanylpentanoic acid, 3-hydroxy-3-methylglutaryl-coenzyme A reductase, GLYCEROL, ... | | 著者 | Steussy, C.N, Stauffacher, C.V, Schmidt, T, Burgner II, J.W, Rodwell, V.W, Wrensford, L.V, Critchelow, C.J, Min, J. | | 登録日 | 2012-11-30 | | 公開日 | 2013-07-17 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | A Novel Role for Coenzyme A during Hydride Transfer in 3-Hydroxy-3-methylglutaryl-coenzyme A Reductase.
Biochemistry, 52, 2013
|
|
