3FRS
 
 | | Structure of human IST1(NTD) (residues 1-189)(p43212) | | 分子名称: | GLYCEROL, Uncharacterized protein KIAA0174 | | 著者 | Schubert, H.L, Hill, C.P, Bajorek, M, Sundquist, W.I. | | 登録日 | 2009-01-08 | | 公開日 | 2009-06-30 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Structural basis for ESCRT-III protein autoinhibition.
Nat.Struct.Mol.Biol., 16, 2009
|
|
6CUP
 
 | | Ras:SOS:Ras in complex with a small molecule activator | | 分子名称: | FORMIC ACID, GLYCEROL, GTPase HRas, ... | | 著者 | Phan, J, Abbott, J, Fesik, S.W. | | 登録日 | 2018-03-26 | | 公開日 | 2019-02-06 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.833 Å) | | 主引用文献 | Discovery of Quinazolines That Activate SOS1-Mediated Nucleotide Exchange on RAS.
ACS Med Chem Lett, 9, 2018
|
|
8IFR
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 3 | | 分子名称: | (1R,2S,5S)-3-[(2S)-2-(tert-butylcarbamoylamino)-3,3-dimethyl-butanoyl]-6,6-dimethyl-N-[(2S)-5-oxidanylidene-1-[(3S)-2-oxidanylidenepyrrolidin-3-yl]hex-3-en-2-yl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | 登録日 | 2023-02-19 | | 公開日 | 2023-10-18 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
8IGX
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 9 (simnotrelvir, SIM0417, SSD8432) | | 分子名称: | (8~{S})-~{N}-[(1~{S})-1-cyano-2-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]ethyl]-7-[(2~{S})-3,3-dimethyl-2-[2,2,2-tris(fluoranyl)ethanoylamino]butanoyl]-1,4-dithia-7-azaspiro[4.4]nonane-8-carboxamide, 3C-like proteinase nsp5 | | 著者 | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | 登録日 | 2023-02-21 | | 公開日 | 2023-10-18 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
5VVT
 
 | | Structural Investigations of the Substrate Specificity of Human O-GlcNAcase | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ELK1 peptide, Protein O-GlcNAcase | | 著者 | Li, B, Jiang, J, Li, H, Hu, C.-W. | | 登録日 | 2017-05-20 | | 公開日 | 2017-09-27 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural insights into the substrate binding adaptability and specificity of human O-GlcNAcase.
Nat Commun, 8, 2017
|
|
8IGY
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | 登録日 | 2023-02-21 | | 公開日 | 2023-10-18 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
5C3S
 
 | | Crystal structure of the full-length Neurospora crassa T7H in complex with alpha-KG and 5-formyluracil (5fU) | | 分子名称: | 1,2-ETHANEDIOL, 2,4-dioxo-1,2,3,4-tetrahydropyrimidine-5-carbaldehyde, 2-OXOGLUTARIC ACID, ... | | 著者 | Li, W, Zhang, T, Ding, J. | | 登録日 | 2015-06-17 | | 公開日 | 2015-10-21 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Molecular basis for the substrate specificity and catalytic mechanism of thymine-7-hydroxylase in fungi
Nucleic Acids Res., 43, 2015
|
|
3JC8
 
 | | Architectural model of the type IVa pilus machine in a piliated state | | 分子名称: | LysM domain protein, PilA, PilN, ... | | 著者 | Chang, Y.-W, Rettberg, L.A, Jensen, G.J. | | 登録日 | 2015-11-24 | | 公開日 | 2016-03-16 | | 最終更新日 | 2018-07-18 | | 実験手法 | ELECTRON MICROSCOPY | | 主引用文献 | Architecture of the type IVa pilus machine.
Science, 351, 2016
|
|
5W53
 
 | |
3G2G
 
 | | S437Y Mutant of human muscle pyruvate kinase, isoform M2 | | 分子名称: | Pyruvate kinase isozymes M1/M2, SULFATE ION, UNKNOWN ATOM OR ION | | 著者 | Hong, B, Dimov, S, Allali-Hassani, A, Tempel, W, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, c, Weigelt, J, Bochkarev, A, Vedadi, M, Park, H, Structural Genomics Consortium (SGC) | | 登録日 | 2009-01-31 | | 公開日 | 2009-03-03 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | S437Y Mutant of human muscle pyruvate kinase, isoform M2
To be Published
|
|
1ZN0
 
 | | Coordinates of RRF and EF-G fitted into Cryo-EM map of the 50S subunit bound with both EF-G (GDPNP) and RRF | | 分子名称: | 16S RIBOSOMAL RNA, ELONGATION FACTOR G, Ribosome recycling factor | | 著者 | Gao, N, Zavialov, A.V, Li, W, Sengupta, J, Valle, M, Gursky, R.P, Ehrenberg, M, Frank, J. | | 登録日 | 2005-05-11 | | 公開日 | 2005-06-14 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (15.5 Å) | | 主引用文献 | Mechanism for the disassembly of the posttermination complex inferred from cryo-EM studies.
Mol.Cell, 18, 2005
|
|
4NN9
 
 | | REFINED ATOMIC STRUCTURES OF N9 SUBTYPE INFLUENZA VIRUS NEURAMINIDASE AND ESCAPE MUTANTS | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NEURAMINIDASE N9, ... | | 著者 | Tulip, W.R, Varghese, J.N, Baker, A.T, Vandonkelaar, A, Laver, W.G, Webster, R.G, Colman, P.M. | | 登録日 | 1991-03-28 | | 公開日 | 1992-07-15 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Refined atomic structures of N9 subtype influenza virus neuraminidase and escape mutants.
J.Mol.Biol., 221, 1991
|
|
3K1E
 
 | | Crystal structure of odorant binding protein 1 (AaegOBP1) from Aedes aegypti | | 分子名称: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Leite, N.R, Krogh, R, Leal, W.S, Iulek, J, Oliva, G. | | 登録日 | 2009-09-27 | | 公開日 | 2009-12-08 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structure of an odorant-binding protein from the mosquito Aedes aegypti suggests a binding pocket covered by a pH-sensitive "Lid".
Plos One, 4, 2009
|
|
1YY6
 
 | | The Crystal Structure of the N-terminal domain of HAUSP/USP7 complexed with an EBNA1 peptide | | 分子名称: | Epstein-Barr nuclear antigen-1, SODIUM ION, Ubiquitin carboxyl-terminal hydrolase 7 | | 著者 | Saridakis, V, Sheng, Y, Sarkari, F, Holowaty, M, Shire, K, Nguyen, T, Zhang, R, Liao, J, Lee, W, Edwards, A.M, Arrowsmith, C.H, Frappier, L. | | 登録日 | 2005-02-23 | | 公開日 | 2005-04-05 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure of the p53 binding domain of HAUSP/USP7 bound to Epstein-Barr nuclear antigen 1 implications for EBV-mediated immortalization.
Mol.Cell, 18, 2005
|
|
8OF7
 
 | | Cyc15 Diels Alderase | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Back, C.R, Barringer, R.W.L, Zorn, K, Manzo-Ruiz, M, Race, P.R. | | 登録日 | 2023-03-14 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Interrogation of an Enzyme Library Reveals the Catalytic Plasticity of Naturally Evolved [4+2] Cyclases.
Chembiochem, 24, 2023
|
|
6X4D
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 5-(cyclopropylmethyl)-7-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-8-methyl-2-naphthonitrile (JLJ678), a Non-nucleoside Inhibitor | | 分子名称: | 5-(cyclopropylmethyl)-7-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-8-methylnaphthalene-2-carbonitrile, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | 著者 | Chan, A.H, Duong, V.N, Ippolito, J.A, Jorgensen, W.L, Anderson, K.S. | | 登録日 | 2020-05-22 | | 公開日 | 2020-07-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structural investigation of 2-naphthyl phenyl ether inhibitors bound to WT and Y181C reverse transcriptase highlights key features of the NNRTI binding site.
Protein Sci., 29, 2020
|
|
5AAC
 
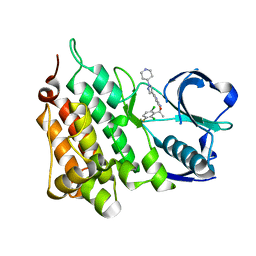 | | Structure of C1156Y Mutant Human Anaplastic Lymphoma Kinase in Complex with Crizotinib | | 分子名称: | 3-[(1R)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]-5-(1-piperidin-4-yl-1H-pyrazol-4-yl)pyridin-2-amine, ALK TYROSINE KINASE RECEPTOR | | 著者 | McTigue, M, Deng, Y, Liu, W, Brooun, A, Stewart, A. | | 登録日 | 2015-07-23 | | 公開日 | 2016-06-08 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Resensitization to Crizotinib by the Lorlatinib Alk Resistance Mutation L1198F.
N.Engl.J.Med., 374, 2016
|
|
4CXW
 
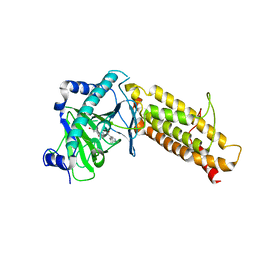 | | Crystal structure of human FTO in complex with subfamily-selective inhibitor 12 | | 分子名称: | (2E)-4-[N'-(4-benzyl-pyridine-3-carbonyl)-hydrazino]-4-oxo-but-2-enoic acid, ALPHA-KETOGLUTARATE-DEPENDENT DIOXYGENASE FTO, NICKEL (II) ION | | 著者 | Toh, D.W, Sun, L, Tan, J, Chen, Y, Lau, L.Z.M, Hong, W, Woon, E.C.Y, Gao, Y.G. | | 登録日 | 2014-04-09 | | 公開日 | 2014-10-01 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | A strategy based on nucleotide specificity leads to a subfamily-selective and cell-active inhibitor ofN6-methyladenosine demethylase FTO.
Chem Sci, 6, 2015
|
|
3DAQ
 
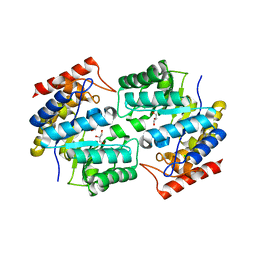 | | Crystal structure of dihydrodipicolinate synthase from methicillin-resistant Staphylococcus aureus | | 分子名称: | CHLORIDE ION, Dihydrodipicolinate synthase, GLYCEROL | | 著者 | Dobson, R.C.J, Burgess, B.R, Jameson, G.B, Gerrard, J.A, Parker, M.W, Perugini, M.A. | | 登録日 | 2008-05-29 | | 公開日 | 2008-08-05 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Structure and evolution of a novel dimeric enzyme from a clinically-important bacterial pathogen.
J.Biol.Chem., 2008
|
|
3DX9
 
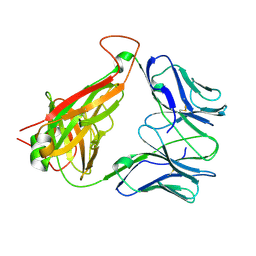 | | Crystal Structure of the DM1 TCR at 2.75A | | 分子名称: | DM1 T cell receptor alpha chain, DM1 T cell receptor beta chain | | 著者 | Archbold, J.K, Macdonald, W.A, Gras, S, Rossjohn, J. | | 登録日 | 2008-07-24 | | 公開日 | 2009-01-27 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Natural micropolymorphism in human leukocyte antigens provides a basis for genetic control of antigen recognition.
J.Exp.Med., 206, 2009
|
|
5WP6
 
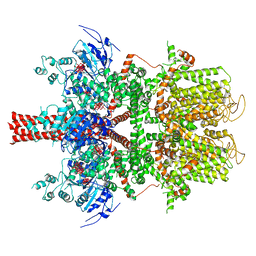 | | Cryo-EM structure of a human TRPM4 channel in complex with calcium and decavanadate | | 分子名称: | DECAVANADATE, Transient receptor potential cation channel subfamily M member 4 | | 著者 | Winkler, P.A, Huang, Y, Sun, W, Du, J, Lu, W. | | 登録日 | 2017-08-03 | | 公開日 | 2017-12-13 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Electron cryo-microscopy structure of a human TRPM4 channel.
Nature, 552, 2017
|
|
5CU1
 
 | |
1YCE
 
 | | Structure of the rotor ring of F-type Na+-ATPase from Ilyobacter tartaricus | | 分子名称: | NONAN-1-OL, SODIUM ION, subunit c | | 著者 | Meier, T, Polzer, P, Diederichs, K, Welte, W, Dimroth, P. | | 登録日 | 2004-12-22 | | 公開日 | 2005-04-12 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure of the rotor ring of F-Type Na+-ATPase from Ilyobacter tartaricus.
Science, 308, 2005
|
|
8W4P
 
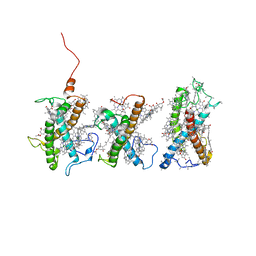 | | Structure of PSII-FCPII-I/J/K complex in the PSII-FCPII supercomplex from Cyclotella meneghiniana | | 分子名称: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, CHLOROPHYLL A, ... | | 著者 | Shen, L.L, Li, Z.H, Shen, J.R, Wang, W.D. | | 登録日 | 2023-08-24 | | 公開日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (3.48 Å) | | 主引用文献 | Structural insights into photosystem II supercomplex and trimeric FCP antennae of a centric diatom Cyclotella meneghiniana.
Nat Commun, 14, 2023
|
|
5GM9
 
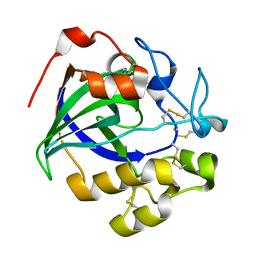 | | Crystal structure of a glycoside hydrolase in complex with cellobiose | | 分子名称: | Glycoside hydrolase family 45 protein, beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Gao, J, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | 登録日 | 2016-07-13 | | 公開日 | 2017-04-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | Characterization and crystal structure of a thermostable glycoside hydrolase family 45 1,4-beta-endoglucanase from Thielavia terrestris
Enzyme Microb. Technol., 99, 2017
|
|
