8HCD
 
 | | Crystal structure of mTREX1-DNA product complex (dNMP) | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, COBALT (II) ION, ... | | Authors: | Hsiao, Y.Y, Huang, K.W, Wu, C.Y. | | Deposit date: | 2022-11-01 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular insight into the specific enzymatic properties of TREX1 revealing the diverse functions in processing RNA and DNA/RNA hybrids.
Nucleic Acids Res., 51, 2023
|
|
6QCN
 
 | | Human Sirt2 in complex with ADP-ribose and the inhibitor quercetin | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, NAD-dependent protein deacetylase sirtuin-2, ZINC ION, ... | | Authors: | Riemer, S, You, W, Steegborn, C. | | Deposit date: | 2018-12-29 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural basis for the activation and inhibition of Sirtuin 6 by quercetin and its derivatives.
Sci Rep, 9, 2019
|
|
8HCE
 
 | | Crystal structure of mTREX1-CMP complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AMMONIUM ION, CYTIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Hsiao, Y.Y, Huang, K.W, Wu, C.Y. | | Deposit date: | 2022-11-01 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Molecular insight into the specific enzymatic properties of TREX1 revealing the diverse functions in processing RNA and DNA/RNA hybrids.
Nucleic Acids Res., 51, 2023
|
|
8HCF
 
 | | Crystal structure of mTREX1-UMP complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SULFATE ION, Three-prime repair exonuclease 1, ... | | Authors: | Hsiao, Y.Y, Huang, K.W, Wu, C.Y. | | Deposit date: | 2022-11-01 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular insight into the specific enzymatic properties of TREX1 revealing the diverse functions in processing RNA and DNA/RNA hybrids.
Nucleic Acids Res., 51, 2023
|
|
6KMA
 
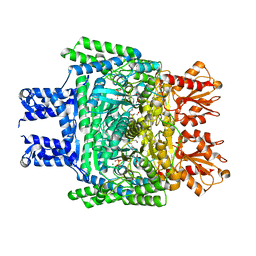 | | Crystal structure of SucA with glycolaldehyde-1-13C from Vibrio vulnificus | | Descriptor: | 2-oxidanylethanal, CALCIUM ION, HEXAETHYLENE GLYCOL, ... | | Authors: | Seo, P.W, Kim, J.S. | | Deposit date: | 2019-07-31 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.282 Å) | | Cite: | Understanding the molecular properties of the E1 subunit (SucA) of alpha-ketoglutarate dehydrogenase complex from Vibrio vulnificus for the enantioselective ligation of acetaldehydes into (R)-acetoin.
Catalysis Science And Technology, 2020
|
|
4UUU
 
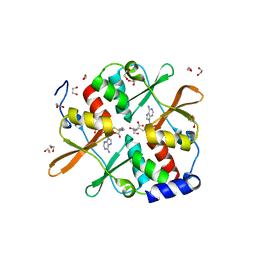 | | 1.7 A resolution structure of human cystathionine beta-synthase regulatory domain (del 516-525) in complex with SAM | | Descriptor: | 1,2-ETHANEDIOL, CYSTATHIONINE BETA-SYNTHASE, S-ADENOSYLMETHIONINE | | Authors: | Kopec, J, McCorvie, T.J, Fitzpatrick, F, Strain-Damerell, C, Froese, D.S, Tallant, C, Burgess-Brown, N, Arrowsmith, C, Edwards, A, Bountra, C, Yue, W.W. | | Deposit date: | 2014-07-31 | | Release date: | 2014-08-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Inter-Domain Communication of Human Cystathionine Beta Synthase: Structural Basis of S-Adenosyl-L-Methionine Activation.
J.Biol.Chem., 289, 2014
|
|
8HQI
 
 | |
1IIN
 
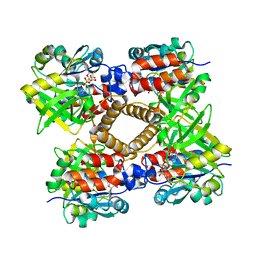 | | thymidylyltransferase complexed with UDP-glucose | | Descriptor: | URIDINE-5'-DIPHOSPHATE-GLUCOSE, glucose-1-phosphate thymidylyltransferase | | Authors: | Barton, W.A, Lesniak, J, Biggins, J.B, Jeffrey, P.D, Jiang, J, Rajashankar, K.R, Thorson, J.S, Nikolov, D.B. | | Deposit date: | 2001-04-23 | | Release date: | 2001-05-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure, mechanism and engineering of a nucleotidylyltransferase as a first step toward glycorandomization.
Nat.Struct.Biol., 8, 2001
|
|
1IND
 
 | |
5H9D
 
 | | Crystal structure of Heptaprenyl Diphosphate Synthase from Staphylococcus aureus | | Descriptor: | C-terminal peptide from Heptaprenyl diphosphate synthase (HEPPP synthase) subunit 1 family protein, Farnesyl pyrophosphate synthetase, Heptaprenyl diphosphate synthase (HEPPP synthase) subunit 1 family protein, ... | | Authors: | Wei, H.L, Liu, W.D, Zheng, Y.Y, Ko, T.P, Chen, C.C, Guo, R.T. | | Deposit date: | 2015-12-28 | | Release date: | 2016-12-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structure, Function, and Inhibition of Staphylococcus aureus Heptaprenyl Diphosphate Synthase
ChemMedChem, 11, 2016
|
|
7YVR
 
 | | Crystal Structure of L-Threonine Aldolase from Neptunomonas marina | | Descriptor: | GLYCEROL, L-threonine aldolase | | Authors: | He, Y.Z, Wang, J, Yan, W.P, Zhang, Y, Feng, Y. | | Deposit date: | 2022-08-19 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and Engineering of the L-Threonine Aldolase from Neptunomonas marine for the Efficient Synthesis of beta-Hydroxy-alpha-amino Acids via C-C Formation
Acs Catalysis, 2023
|
|
7YL6
 
 | | Cell surface protein YwfG protein complexed with alpha-1,2-mannobiose | | Descriptor: | CALCIUM ION, GRAM_POS_ANCHORING domain-containing protein, SULFATE ION, ... | | Authors: | Tsuchiya, W, Fujimoto, Z, Suzuki, C. | | Deposit date: | 2022-07-25 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Cell-surface protein YwfG of Lactococcus lactis binds to alpha-1,2-linked mannose.
Plos One, 18, 2023
|
|
7YL4
 
 | | Cell surface protein YwfG protein (apo form) | | Descriptor: | CALCIUM ION, GRAM_POS_ANCHORING domain-containing protein, SULFATE ION | | Authors: | Tsuchiya, W, Fujimoto, Z, Suzuki, C. | | Deposit date: | 2022-07-25 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cell-surface protein YwfG of Lactococcus lactis binds to alpha-1,2-linked mannose.
Plos One, 18, 2023
|
|
5HCN
 
 | | GPN-loop GTPase Npa3 in complex with GMPPCP | | Descriptor: | GLYCEROL, GPN-loop GTPase 1, LAURIC ACID, ... | | Authors: | Niesser, J, Wagner, F.R, Kostrewa, D, Muehlbacher, W, Cramer, P. | | Deposit date: | 2016-01-04 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of GPN-Loop GTPase Npa3 and Implications for RNA Polymerase II Assembly.
Mol.Cell.Biol., 36, 2015
|
|
7SD4
 
 | | SARS-CoV-2 Nucleocapsid N-terminal domain (N-NTD) protein | | Descriptor: | Nucleoprotein | | Authors: | Sarkar, S, Runge, B, Russell, R.W, Calero, D, Zeinalilathori, S, Quinn, C.M, Lu, M, Calero, G, Gronenborn, A.M, Polenova, T. | | Deposit date: | 2021-09-29 | | Release date: | 2022-06-08 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Atomic-Resolution Structure of SARS-CoV-2 Nucleocapsid Protein N-Terminal Domain.
J.Am.Chem.Soc., 144, 2022
|
|
6JQ7
 
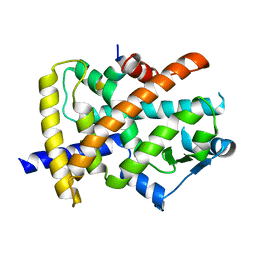 | |
5HI1
 
 | | Backbone Modifications in the Protein GB1 Helix: Aib24, beta-3-Lys28, beta-3-Lys31, Aib35 | | Descriptor: | ACETATE ION, Immunoglobulin G-binding protein G | | Authors: | Tavenor, N.A, Reinert, Z.E, Lengyel, G.A, Griffith, B.D, Horne, W.S. | | Deposit date: | 2016-01-11 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Comparison of design strategies for alpha-helix backbone modification in a protein tertiary fold.
Chem.Commun.(Camb.), 52, 2016
|
|
7YTH
 
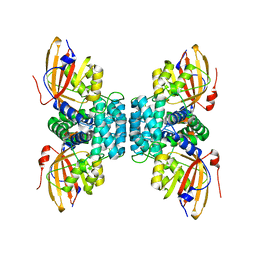 | | Structure of OCPx1 from Nostoc flagelliforme CCNUN1 | | Descriptor: | Ketosteroid isomerase-related protein | | Authors: | Yang, Y.W, Liu, K, Chen, S.Z, Chen, M, Qiu, B.S. | | Deposit date: | 2022-08-14 | | Release date: | 2023-06-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Functional specialization of expanded orange carotenoid protein paralogs in subaerial Nostoc species.
Plant Physiol., 192, 2023
|
|
7YRY
 
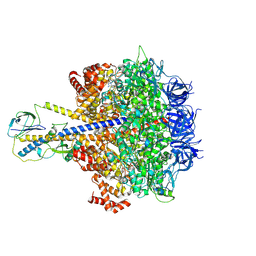 | | F1-ATPase of Acinetobacter baumannii | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Saw, W.-G, Grueber, G. | | Deposit date: | 2022-08-11 | | Release date: | 2023-06-21 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Atomic insights of an up and down conformation of the Acinetobacter baumannii F 1 -ATPase subunit epsilon and deciphering the residues critical for ATP hydrolysis inhibition and ATP synthesis.
Faseb J., 37, 2023
|
|
1ITF
 
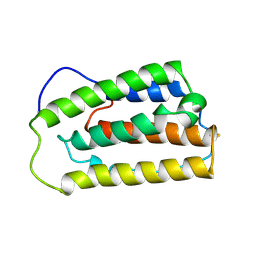 | | INTERFERON ALPHA-2A, NMR, 24 STRUCTURES | | Descriptor: | INTERFERON ALPHA-2A | | Authors: | Klaus, W, Gsell, B, Labhardt, A.M, Wipf, B, Senn, H. | | Deposit date: | 1997-08-22 | | Release date: | 1997-12-03 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional high resolution structure of human interferon alpha-2a determined by heteronuclear NMR spectroscopy in solution.
J.Mol.Biol., 274, 1997
|
|
1J2Z
 
 | | Crystal structure of UDP-N-acetylglucosamine acyltransferase | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, L(+)-TARTARIC ACID, SULFATE ION, ... | | Authors: | Lee, B.I, Suh, S.W. | | Deposit date: | 2003-01-15 | | Release date: | 2004-01-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of UDP-N-acetylglucosamine acyltransferase from Helicobacter pylori
Proteins, 53, 2003
|
|
5HKN
 
 | | Crystal structure de novo designed fullerene organizing protein complex with fullerene | | Descriptor: | (C_{60}-I_{h})[5,6]fullerene, fullerene organizing protein | | Authors: | Paul, J, Acharya, R, Kim, K.-H, Kim, Y.H, Kim, N.H, Grigoryan, G, DeGardo, W.F. | | Deposit date: | 2016-01-14 | | Release date: | 2016-05-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.761 Å) | | Cite: | Protein-directed self-assembly of a fullerene crystal.
Nat Commun, 7, 2016
|
|
7YPF
 
 | |
1IDC
 
 | | ISOCITRATE DEHYDROGENASE FROM E.COLI (MUTANT K230M), STEADY-STATE INTERMEDIATE COMPLEX DETERMINED BY LAUE CRYSTALLOGRAPHY | | Descriptor: | 2-OXALOSUCCINIC ACID, ISOCITRATE DEHYDROGENASE, MAGNESIUM ION | | Authors: | Bolduc, J.M, Dyer, D.H, Scott, W.G, Singer, P, Sweet, R.M, Koshland Junior, D.E, Stoddard, B.L. | | Deposit date: | 1995-01-18 | | Release date: | 1996-03-08 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutagenesis and Laue structures of enzyme intermediates: isocitrate dehydrogenase.
Science, 268, 1995
|
|
5HSG
 
 | | Crystal structure of an ABC transporter Solute Binding Protein from Klebsiella pneumoniae (KPN_01730, target EFI-511059), APO open structure | | Descriptor: | MAGNESIUM ION, Putative ABC transporter, nucleotide binding/ATPase protein | | Authors: | Roth, Y, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2016-01-25 | | Release date: | 2016-03-02 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of an ABC transporter Solute Binding Protein from Klebsiella pneumoniae (KPN_01730, target EFI-511059), APO open structure
To be published
|
|
