4W8T
 
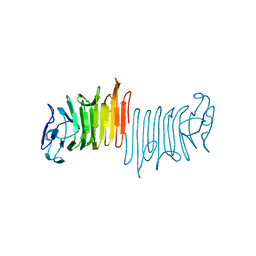 | |
4WB0
 
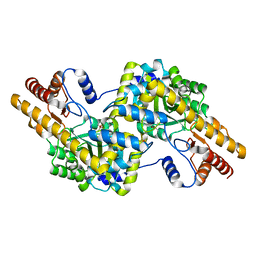 | |
6K0S
 
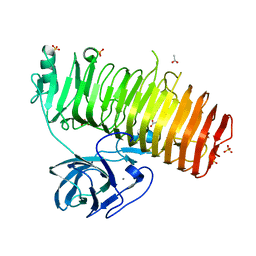 | | Catalytic domain of GH87 alpha-1,3-glucanase D1069A in complex with nigerose | | Descriptor: | ACETIC ACID, Alpha-1,3-glucanase, CALCIUM ION, ... | | Authors: | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.534 Å) | | Cite: | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
6K1H
 
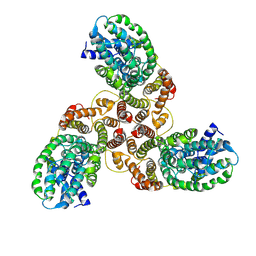 | | Structure of membrane protein | | Descriptor: | PTS mannose transporter subunit IID, PTS system mannose-specific EIIC component, alpha-D-mannopyranose | | Authors: | Wang, J.W, Zeng, J.W. | | Deposit date: | 2019-05-10 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structure of the mannose transporter of the bacterial phosphotransferase system.
Cell Res., 29, 2019
|
|
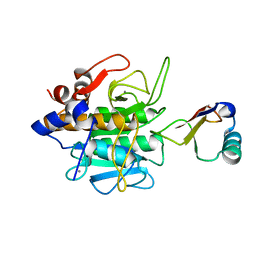
1CSE
 
 | |
4UV3
 
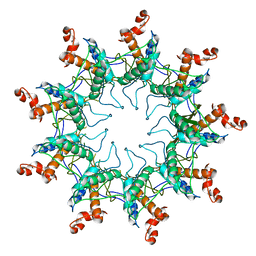 | | Structure of the curli transport lipoprotein CsgG in its membrane- bound conformation | | Descriptor: | CURLI PRODUCTION ASSEMBLY/TRANSPORT COMPONENT CSGG | | Authors: | Goyal, P, Krasteva, P.V, Gerven, N.V, Gubellini, F, Broeck, I.V.D, Troupiotis-Tsailaki, A, Jonckheere, W, Pehau-Arnaudet, G, Pinkner, J.S, Chapman, M.R, Hultgren, S.J, Howorka, S, Fronzes, R, Remaut, H. | | Deposit date: | 2014-08-04 | | Release date: | 2014-09-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Structural and Mechanistic Insights Into the Bacterial Amyloid Secretion Channel Csgg.
Nature, 516, 2014
|
|
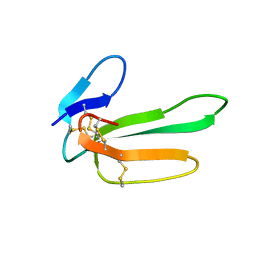
1CVO
 
 | |
6KBY
 
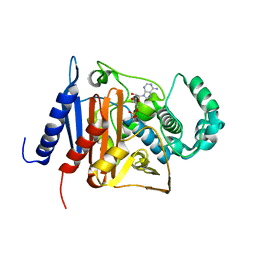 | | Crystal structure of a class C beta lactamase in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Beta-lactamase | | Authors: | Bae, D.W, Jung, Y.E, An, Y.J, Na, J.H, Cha, S.S. | | Deposit date: | 2019-06-26 | | Release date: | 2019-10-16 | | Last modified: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (1.097 Å) | | Cite: | Structural Insights into Catalytic Relevances of Substrate Poses in ACC-1.
Antimicrob.Agents Chemother., 63, 2019
|
|
6KJJ
 
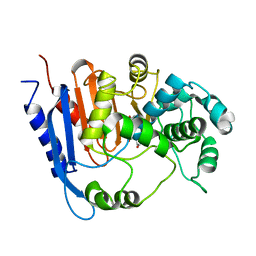 | | Functional and structural insights into the unusual oxyanion hole-like geometry in macrolactin acyltransferase selective for dicarboxylic acyl donors | | Descriptor: | 4-(2-acetamidoethylsulfanyl)-4-oxidanylidene-butanoic acid, Putative beta-lactamase | | Authors: | Xiao, F, Dong, S, Feng, Y, Li, W. | | Deposit date: | 2019-07-22 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.492 Å) | | Cite: | Structural Basis of Specificity for Carboxyl-Terminated Acyl Donors in a Bacterial Acyltransferase.
J.Am.Chem.Soc., 142, 2020
|
|
6KKG
 
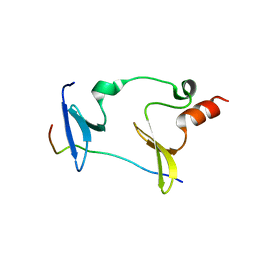 | | Crystal structure of MAGI2-Dendrin complex | | Descriptor: | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2, Peptide from Dendrin | | Authors: | Zhu, J.W, Zhang, H.J, Lin, L, Zhang, R.G. | | Deposit date: | 2019-07-25 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Phase separation of MAGI2-mediated complex underlies formation of slit diaphragm complex in glomerular filtration barrier
To Be Published
|
|
1D4U
 
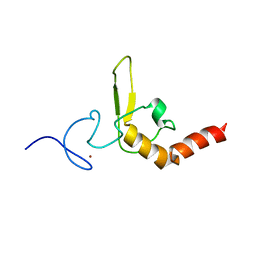 | | INTERACTIONS OF HUMAN NUCLEOTIDE EXCISION REPAIR PROTEIN XPA WITH RPA70 AND DNA: CHEMICAL SHIFT MAPPING AND 15N NMR RELAXATION STUDIES | | Descriptor: | NUCLEOTIDE EXCISION REPAIR PROTEIN XPA (XPA-MBD), ZINC ION | | Authors: | Buchko, G.W, Daughdrill, G.W, de Lorimier, R, Rao, S, Isern, N.G, Lingbeck, J, Taylor, J, Wold, M.S, Gochin, M, Spicer, L.D, Lowry, D.F, Kennedy, M.A. | | Deposit date: | 1999-10-06 | | Release date: | 1999-10-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Interactions of human nucleotide excision repair protein XPA with DNA and RPA70 Delta C327: chemical shift mapping and 15N NMR relaxation studies.
Biochemistry, 38, 1999
|
|
6KEA
 
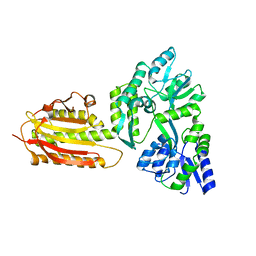 | | crystal structure of MBP-tagged REV7-IpaB complex | | Descriptor: | Maltose-binding periplasmic protein,LINKER,hREV7,LINKER,Invasin IpaB,hREV3 | | Authors: | Wang, X, Pernicone, N, Pertz, L, Hua, D.P, Zhang, T.Q, Listovsky, T, Xie, W. | | Deposit date: | 2019-07-04 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | REV7 has a dynamic adaptor region to accommodate small GTPase RAN/ShigellaIpaB ligands, and its activity is regulated by the RanGTP/GDP switch.
J.Biol.Chem., 294, 2019
|
|
6KFU
 
 | |
4W7H
 
 | | Crystal Structure of DEH Reductase A1-R Mutant | | Descriptor: | Carbonyl reductase | | Authors: | Takase, R, Mikami, B, Kawai, S, Murata, K, Hashimoto, W. | | Deposit date: | 2014-08-22 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structure-based Conversion of the Coenzyme Requirement of a Short-chain Dehydrogenase/Reductase Involved in Bacterial Alginate Metabolism.
J.Biol.Chem., 289, 2014
|
|
4W8J
 
 | | Structure of the full-length insecticidal protein Cry1Ac reveals intriguing details of toxin packaging into in vivo formed crystals | | Descriptor: | CALCIUM ION, POTASSIUM ION, Pesticidal crystal protein cry1Ac | | Authors: | Evdokimov, A.G, Moshiri, F, Sturman, E.J, Rydel, T.J, Zheng, M, Seale, J.W, Franklin, S. | | Deposit date: | 2014-08-25 | | Release date: | 2014-09-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structure of the full-length insecticidal protein Cry1Ac reveals intriguing details of toxin packaging into in vivo formed crystals.
Protein Sci., 23, 2014
|
|
1CXL
 
 | |
4URD
 
 | | Cryo-EM map of Trigger Factor bound to a translating ribosome | | Descriptor: | TRIGGER FACTOR | | Authors: | Deeng, J, Chan, K.Y, van der Sluis, E, Bischoff, L, Berninghausen, O, Han, W, Gumbart, J, Schulten, K, Beatrix, B, Beckmann, R. | | Deposit date: | 2014-06-27 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Dynamic Behavior of Trigger Factor on the Ribosome.
J.Mol.Biol., 428, 2016
|
|
1D3C
 
 | | MICHAELIS COMPLEX OF BACILLUS CIRCULANS STRAIN 251 CYCLODEXTRIN GLYCOSYLTRANSFERASE WITH GAMMA-CYCLODEXTRIN | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, CYCLODEXTRIN GLYCOSYLTRANSFERASE, ... | | Authors: | Uitdehaag, J.C.M, Kalk, K.H, van der Veen, B.A, Dijkhuizen, L, Dijkstra, B.W. | | Deposit date: | 1999-09-29 | | Release date: | 1999-12-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The cyclization mechanism of cyclodextrin glycosyltransferase (CGTase) as revealed by a gamma-cyclodextrin-CGTase complex at 1.8-A resolution.
J.Biol.Chem., 274, 1999
|
|
6K68
 
 | | Application of anti-helix antibodies in protein structure determination (8420-3MNZ) | | Descriptor: | 3MNZ Variable heavy chain, 3MNZ Variable light chain, Protein A | | Authors: | Lee, J.O, Jin, M.S, Kim, J.W, Kim, S, Lee, H, Cho, G.Y. | | Deposit date: | 2019-06-01 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Application of antihelix antibodies in protein structure determination.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5TMN
 
 | | Slow-and fast-binding inhibitors of thermolysin display different modes of binding. crystallographic analysis of extended phosphonamidate transition-state analogues | | Descriptor: | CALCIUM ION, N-[(S)-({[(benzyloxy)carbonyl]amino}methyl)(hydroxy)phosphoryl]-L-leucyl-L-leucine, THERMOLYSIN, ... | | Authors: | Holden, H.M, Tronrud, D.E, Monzingo, A.F, Weaver, L.H, Matthews, B.W. | | Deposit date: | 1987-06-29 | | Release date: | 1989-01-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Slow- and fast-binding inhibitors of thermolysin display different modes of binding: crystallographic analysis of extended phosphonamidate transition-state analogues.
Biochemistry, 26, 1987
|
|
1CIN
 
 | | THE POSITIONS OF HIS-64 AND A BOUND WATER IN HUMAN CARBONIC ANHYDRASE II UPON BINDING THREE STRUCTURALLY RELATED INHIBITORS | | Descriptor: | (4S-TRANS)-4-(METHYLAMINO)-5,6-DIHYDRO-6-METHYL-4H-THIENO(2,3-B)THIOPYRAN-2-SULFONAMIDE-7,7-DIOXIDE, CARBONIC ANHYDRASE II, METHYL MERCURY ION, ... | | Authors: | Smith, G.M, Alexander, R.S, Christianson, D.W, Mckeever, B.M, Ponticello, G.S, Springer, J.P, Randall, W.C, Baldwin, J.J, Habecker, C.N. | | Deposit date: | 1993-10-20 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Positions of His-64 and a bound water in human carbonic anhydrase II upon binding three structurally related inhibitors.
Protein Sci., 3, 1994
|
|
1CIL
 
 | | THE POSITIONS OF HIS-64 AND A BOUND WATER IN HUMAN CARBONIC ANHYDRASE II UPON BINDING THREE STRUCTURALLY RELATED INHIBITORS | | Descriptor: | (4S-TRANS)-4-(ETHYLAMINO)-5,6-DIHYDRO-6-METHYL-4H-THIENO(2,3-B)THIOPYRAN-2-SULFONAMIDE-7,7-DIOXIDE, CARBONIC ANHYDRASE II, ZINC ION | | Authors: | Smith, G.M, Alexander, R.S, Christianson, D.W, Mckeever, B.M, Ponticello, G.S, Springer, J.P, Randall, W.C, Baldwin, J.J, Habecker, C.N. | | Deposit date: | 1993-10-20 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Positions of His-64 and a bound water in human carbonic anhydrase II upon binding three structurally related inhibitors.
Protein Sci., 3, 1994
|
|
7OV3
 
 | | Crystal structure of pig purple acid phosphatase in complex with Maybridge fragment CC063346, dimethyl sulfoxide and citrate | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Feder, D, McGeary, R.P, Guddat, L.W, Schenk, G. | | Deposit date: | 2021-06-14 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rational Design of Potent Inhibitors of a Metallohydrolase Using a Fragment-Based Approach.
Chemmedchem, 16, 2021
|
|
7OV2
 
 | | Crystal structure of pig purple acid phosphatase in complex with L-glutamine, (poly)ethylene glycol fragments and glycerol | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, FE (III) ION, ... | | Authors: | Feder, D, McGeary, R.P, Guddat, L.W, Schenk, G. | | Deposit date: | 2021-06-14 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Rational Design of Potent Inhibitors of a Metallohydrolase Using a Fragment-Based Approach.
Chemmedchem, 16, 2021
|
|
1DA9
 
 | | ANTHRACYCLINE-DNA INTERACTIONS AT UNFAVOURABLE BASE BASE-PAIR TRIPLET-BINDING SITES: STRUCTURES OF D(CGGCCG)/DAUNOMYCIN AND D(TGGCCA)/ADRIAMYCIN COMPL | | Descriptor: | DNA (5'-D(*TP*GP*GP*CP*CP*A)-3'), DOXORUBICIN | | Authors: | Leonard, G.A, Hambley, T.W, McAuley-Hecht, K, Brown, T, Hunter, W.N. | | Deposit date: | 1993-01-21 | | Release date: | 1993-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Anthracycline-DNA interactions at unfavourable base-pair triplet-binding sites: structures of d(CGGCCG)/daunomycin and d(TGGCCA)/adriamycin complexes.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
