1D5Q
 
 | | SOLUTION STRUCTURE OF A MINI-PROTEIN REPRODUCING THE CORE OF THE CD4 SURFACE INTERACTING WITH THE HIV-1 ENVELOPE GLYCOPROTEIN | | Descriptor: | CHIMERIC MINI-PROTEIN | | Authors: | Vita, C, Drakopoulou, E, Vizzanova, J, Rochette, S, Martin, L, Menez, A, Roumestand, C, Yang, Y.S, Ylisastigui, L, Benjouad, A, Gluckman, J.C. | | Deposit date: | 1999-10-11 | | Release date: | 2000-10-11 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Rational engineering of a miniprotein that reproduces the core of the CD4 site interacting with HIV-1 envelope glycoprotein.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
4BGG
 
 | | Crystal structure of the ACVR1 kinase in complex with LDN-213844 | | Descriptor: | 1,2-ETHANEDIOL, 1-{4-[5-(3,4,5-trimethoxyphenyl)pyridin-3-yl]phenyl}piperazine, ACTIVIN RECEPTOR TYPE-1, ... | | Authors: | Sanvitale, C, Canning, P, Cooper, C, Wang, Y, Mohedas, A.H, Choi, S, Yu, P.B, Cuny, G.D, Nowak, R, Coutandin, D, Vollmar, M, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-03-26 | | Release date: | 2013-04-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structure-Activity Relationship of 3,5-Diaryl-2-Aminopyridine Alk2 Inhibitors Reveals Unaltered Binding Affinity for Fibrodysplasia Ossificans Progressiva Causing Mutants.
J.Med.Chem., 57, 2014
|
|
1CMR
 
 | | NMR SOLUTION STRUCTURE OF A CHIMERIC PROTEIN, DESIGNED BY TRANSFERRING A FUNCTIONAL SNAKE BETA-HAIRPIN INTO A SCORPION ALPHA/BETA SCAFFOLD (PH 3.5, 20C), NMR, 18 STRUCTURES | | Descriptor: | CHARYBDOTOXIN, ALPHA CHIMERA | | Authors: | Zinn-Justin, S, Guenneugues, M, Drakopoulou, E, Gilquin, B, Vita, C, Menez, A. | | Deposit date: | 1996-03-15 | | Release date: | 1996-08-01 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Transfer of a beta-hairpin from the functional site of snake curaremimetic toxins to the alpha/beta scaffold of scorpion toxins: three-dimensional solution structure of the chimeric protein.
Biochemistry, 35, 1996
|
|
1BAH
 
 | | A TWO DISULFIDE DERIVATIVE OF CHARYBDOTOXIN WITH DISULFIDE 13-33 REPLACED BY TWO ALPHA-AMINOBUTYRIC ACIDS, NMR, 30 STRUCTURES | | Descriptor: | CHARYBDOTOXIN | | Authors: | Song, J, Gilquin, B, Jamin, N, Guenneugues, M, Dauplais, M, Vita, C, Menez, A. | | Deposit date: | 1996-06-06 | | Release date: | 1997-01-11 | | Last modified: | 2020-01-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of a two-disulfide derivative of charybdotoxin: structural evidence for conservation of scorpion toxin alpha/beta motif and its hydrophobic side chain packing.
Biochemistry, 36, 1997
|
|
1YYL
 
 | | crystal structure of CD4M33, a scorpion-toxin mimic of CD4, in complex with HIV-1 YU2 gp120 envelope glycoprotein and anti-HIV-1 antibody 17b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CD4M33, scorpion-toxin mimic of CD4, ... | | Authors: | Huang, C.C, Stricher, F, Martin, L, Decker, J.M, Majeed, S, Barthe, P, Hendrickson, W.A, Robinson, J, Roumestand, C, Sodroski, J, Wyatt, R, Shaw, G.M, Vita, C, Kwong, P.D. | | Deposit date: | 2005-02-25 | | Release date: | 2005-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Scorpion-toxin mimics of CD4 in complex with human immunodeficiency virus gp120 crystal structures, molecular mimicry, and neutralization breadth.
Structure, 13, 2005
|
|
1YYM
 
 | | crystal structure of F23, a scorpion-toxin mimic of CD4, in complex with HIV-1 YU2 gp120 envelope glycoprotein and anti-HIV-1 antibody 17b | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Exterior membrane glycoprotein(GP120),Exterior membrane glycoprotein(GP120),Exterior membrane glycoprotein(GP120), ... | | Authors: | Huang, C.C, Stricher, F, Martin, L, Decker, J.M, Majeed, S, Barthe, P, Hendrickson, W.A, Robinson, J, Roumestand, C, Sodroski, J, Wyatt, R, Shaw, G.M, Vita, C, Kwong, P.D. | | Deposit date: | 2005-02-25 | | Release date: | 2005-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Scorpion-toxin mimics of CD4 in complex with human immunodeficiency virus gp120 crystal structures, molecular mimicry, and neutralization breadth.
Structure, 13, 2005
|
|
1BGK
 
 | | SEA ANEMONE TOXIN (BGK) WITH HIGH AFFINITY FOR VOLTAGE DEPENDENT POTASSIUM CHANNEL, NMR, 15 STRUCTURES | | Descriptor: | BGK | | Authors: | Dauplais, M, Lecoq, A, Song, J, Cotton, J, Jamin, N, Gilquin, B, Roumestand, C, Vita, C, Harvey, A, Menez, A. | | Deposit date: | 1996-05-08 | | Release date: | 1997-01-27 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | On the convergent evolution of animal toxins. Conservation of a diad of functional residues in potassium channel-blocking toxins with unrelated structures.
J.Biol.Chem., 272, 1997
|
|
1EI0
 
 | |
1RHH
 
 | | Crystal Structure of the Broadly HIV-1 Neutralizing Fab X5 at 1.90 Angstrom Resolution | | Descriptor: | Fab X5, heavy chain, light chain | | Authors: | Darbha, R, Phogat, S, Labrijn, A.F, Shu, Y, Gu, Y, Andrykovitch, M, Zhang, M.Y, Pantophlet, R, Martin, L, Vita, C, Burton, D.R, Dimitrov, D.S, Ji, X. | | Deposit date: | 2003-11-14 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Broadly Cross-Reactive HIV-1-Neutralizing Fab X5 and Fine Mapping of Its Epitope
Biochemistry, 43, 2004
|
|
4URJ
 
 | | Crystal structure of human BJ-TSA-9 | | Descriptor: | 1,2-ETHANEDIOL, PROTEIN FAM83A | | Authors: | Pinkas, D.M, Sanvitale, C, Wang, D, Krojer, T, Kopec, J, Chaikuad, A, Dixon Clarke, S, Berridge, G, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Bullock, A. | | Deposit date: | 2014-06-30 | | Release date: | 2014-10-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Crystal Structure of Human Bj-Tsa-9
To be Published
|
|
6EIX
 
 | | Crystal structure of the kinase domain of the Q207E mutant of ACVR1 (ALK2) in complex with a 2-aminopyridine inhibitor K02288 | | Descriptor: | 1,2-ETHANEDIOL, 3-[6-amino-5-(3,4,5-trimethoxyphenyl)pyridin-3-yl]phenol, Activin receptor type-1 | | Authors: | Williams, E.P, Canning, P, Sanvitale, C.E, Krojer, T, Allerston, C.K, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2017-09-19 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the kinase domain of the Q207E mutant of ACVR1 (ALK2) in complex with K02288
To be published
|
|
4UIJ
 
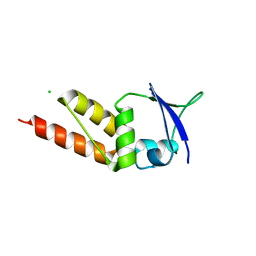 | | Crystal structure of the BTB domain of KCTD13 | | Descriptor: | BTB/POZ DOMAIN-CONTAINING ADAPTER FOR CUL3-MEDIATED RHOA DEGRADATION PROTEIN 1, CHLORIDE ION | | Authors: | Pinkas, D.M, Sanvitale, C.E, Sorell, F.J, Solcan, N, Goubin, S, Canning, P, Williams, E, Chaikuad, A, Dixon Clarke, S.E, Tallant, C, Fonseca, M, Chalk, R, Doutch, J, Krojer, T, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2015-03-30 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural complexity in the KCTD family of Cullin3-dependent E3 ubiquitin ligases.
Biochem. J., 474, 2017
|
|
4UUC
 
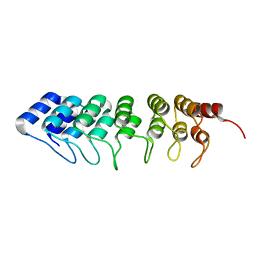 | | Crystal structure of human ASB11 ankyrin repeat domain | | Descriptor: | ANKYRIN REPEAT AND SOCS BOX PROTEIN 11 | | Authors: | Pinkas, D.M, Sanvitale, C, Kragh Nielsen, T, Guo, K, Sorrell, F, Berridge, G, Ayinampudi, V, Wang, D, Newman, J.A, Tallant, C, Chaikuad, A, Canning, P, Kopec, J, Krojer, T, Vollmar, M, Allerston, C.K, Chalk, R, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Bullock, A. | | Deposit date: | 2014-07-25 | | Release date: | 2014-08-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Human Asb11 Ankyrin Repeat Domain
To be Published
|
|
4UYI
 
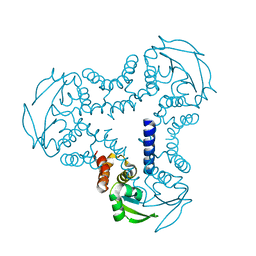 | | Crystal structure of the BTB domain of human SLX4 (BTBD12) | | Descriptor: | STRUCTURE-SPECIFIC ENDONUCLEASE SUBUNIT SLX4 | | Authors: | Pinkas, D.M, Sanvitale, C.E, Strain-Damerell, C, Fairhead, M, Wang, D, Tallant, C, Cooper, C.D.O, Sorrell, F.J, Kopec, J, Chaikuad, A, Fitzpatrick, F, Pike, A.C.W, Hozjan, V, Ying, Z, Roos, A.K, Savitsky, P, Bradley, A, Nowak, R, Filippakopoulos, P, Krojer, T, Burgess-Brown, N.A, Marsden, B.D, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2014-09-01 | | Release date: | 2014-10-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal Structure of the Btb Domain of Human Slx4 (Btbd12)
To be Published
|
|
3T7L
 
 | | Crystal structure of the FYVE domain of endofin (ZFYVE16) at 1.1A resolution | | Descriptor: | 1,2-ETHANEDIOL, ZINC ION, Zinc finger FYVE domain-containing protein 16 | | Authors: | Chaikuad, A, Williams, E, Guo, K, Sanvitale, C, Berridge, G, Krojer, T, Muniz, J.R.C, Canning, P, Phillips, C, Shrestha, A, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-07-30 | | Release date: | 2011-08-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Crystal structure of the FYVE domain of endofin (ZFYVE16) at 1.1A resolution
To be Published
|
|
3SOC
 
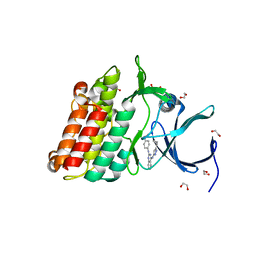 | | Crystal structure of Activin receptor type-IIA (ACVR2A) kinase domain in complex with a quinazolin | | Descriptor: | 1,2-ETHANEDIOL, Activin receptor type-2A, [4-({4-[(5-CYCLOPROPYL-1H-PYRAZOL-3-YL)AMINO]QUINAZOLIN-2-YL}IMINO)CYCLOHEXA-2,5-DIEN-1-YL]ACETONITRILE | | Authors: | Chaikuad, A, Williams, E, Mahajan, P, Cooper, C.D.O, Sanvitale, C, Vollmar, M, Muniz, J.R.C, Yue, W.W, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-06-30 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of Activin receptor type-IIA (ACVR2A) kinase domain in complex with a quinazolin
To be Published
|
|
5A15
 
 | | Crystal structure of the BTB domain of human KCTD16 | | Descriptor: | BTB/POZ DOMAIN-CONTAINING PROTEIN KCTD16 | | Authors: | Pinkas, D.M, Sanvitale, C.E, Solcan, N, Goubin, S, Canning, P, Dixon Clarke, S.E, Talon, R, Wiggers, H.J, Fitzpatrick, F, Tallant, C, Kopec, J, Chalk, R, Doutch, J, Krojer, T, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2015-04-28 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural complexity in the KCTD family of Cullin3-dependent E3 ubiquitin ligases.
Biochem. J., 474, 2017
|
|
5A6R
 
 | | Crystal structure of the BTB domain of human KCTD17 | | Descriptor: | BTB/POZ DOMAIN-CONTAINING PROTEIN KCTD17 | | Authors: | Pinkas, D.M, Sorrell, F, Sanvitale, C.E, Goubin, S, Williams, E, Newman, J.A, Pearce, N.M, Neshich, I, Pike, A.C.W, MacKenzie, A, Quigley, A, Faust, B, Carpenter, E.P, Tallant, C, Kopec, J, Chalk, R, Krojer, T, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2015-06-30 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural complexity in the KCTD family of Cullin3-dependent E3 ubiquitin ligases.
Biochem. J., 474, 2017
|
|
5FTA
 
 | | Crystal structure of the N-terminal BTB domain of human KCTD10 | | Descriptor: | BTB/POZ DOMAIN-CONTAINING ADAPTER FOR CUL3-MEDIATED RHOA DEGRADATION PROTEIN 3, MERCURY (II) ION | | Authors: | Pinkas, D.M, Sanvitale, C.E, Solcan, N, Goubin, S, Tallant, C, Newman, J.A, Kopec, J, Fitzpatrick, F, Talon, R, Collins, P, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2016-01-12 | | Release date: | 2016-02-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural complexity in the KCTD family of Cullin3-dependent E3 ubiquitin ligases.
Biochem. J., 474, 2017
|
|
5G3Q
 
 | | Crystal structure of a hypothetical domain in WNK1 | | Descriptor: | WNK1 | | Authors: | Pinkas, D.M, Bufton, J.C, Sanvitale, C.E, Bartual, S.G, Adamson, R.J, Krojer, T, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2016-04-29 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.613 Å) | | Cite: | Crystal Structure of a Hypothetical Domain in Wnk1
To be Published
|
|
