1U9O
 
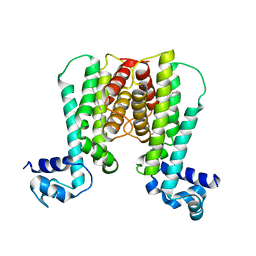 | | Crystal structure of the transcriptional regulator EthR in a ligand bound conformation | | Descriptor: | HEXADECYL OCTANOATE, Transcriptional repressor EthR | | Authors: | Frenois, F, Engohang-Ndong, J, Locht, C, Baulard, A.R, Villeret, V. | | Deposit date: | 2004-08-10 | | Release date: | 2004-11-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of EthR in a Ligand Bound Conformation Reveals Therapeutic Perspectives against Tuberculosis
Mol.Cell, 16, 2004
|
|
7NTT
 
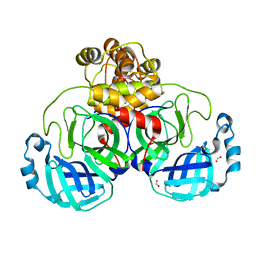 | |
7NTW
 
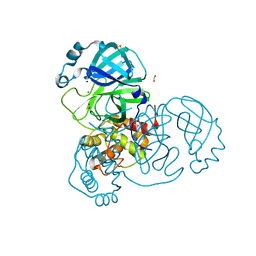 | |
1RWR
 
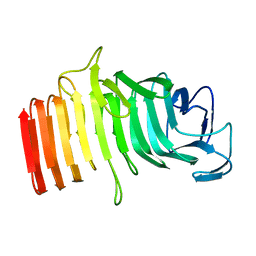 | |
3Q0W
 
 | | ETHR From mycobacterium tuberculosis in complex with compound BDM33066 | | Descriptor: | (2S)-2-amino-3-methyl-1-{4-[3-(thiophen-2-yl)-1,2,4-oxadiazol-5-yl]piperidin-1-yl}butan-1-one, GLYCEROL, HTH-type transcriptional regulator EthR | | Authors: | Flipo, M, Desrose, M, Dirie, B, Carette, X, Leroux, F, Lens, Z, Rucktooa, P, Piveteau, C, Demirkaya, F, Locht, C, Villeret, V, Christophe, T, Jeon, H.K, Brodin, P, Deprez, B, Baulard, A, Willand, N. | | Deposit date: | 2010-12-16 | | Release date: | 2011-12-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural activation of the transcriptional repressor EthR from Mycobacterium tuberculosis by single amino acid change mimicking natural and synthetic ligands.
Nucleic Acids Res., 40, 2012
|
|
3Q0V
 
 | | ETHR From mycobacterium tuberculosis in complex with compound bdm31369 | | Descriptor: | 4-methyl-1-{4-[3-(thiophen-2-yl)-1,2,4-oxadiazol-5-yl]piperidin-1-yl}pentan-1-one, HTH-type transcriptional regulator EthR | | Authors: | Flipo, M, Desroses, M, Dirie, B, Carette, X, Leroux, F, Lens, Z, Rucktooa, P, Leroux, F, Piveteau, C, Demirkaya, F, Locht, C, Villeret, V, Christophe, T, Jeon, H.K, Brodin, P, Deprez, B, Baulard, A, Willand, N. | | Deposit date: | 2010-12-16 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design, synthesis and optimization of new EthR inhibitors. A new alternative approach to fight tuberculosis by boosting ethionamide
To be Published
|
|
3O8H
 
 | | EthR from Mycobacterium tuberculosis in complex with compound BDM14950 | | Descriptor: | 4-iodo-N-[(1-{2-oxo-2-[4-(3-thiophen-2-yl-1,2,4-oxadiazol-5-yl)piperidin-1-yl]ethyl}-1H-1,2,3-triazol-4-yl)methyl]benzenesulfonamide, Transcriptional Regulatory Repressor protein (TETR-Family) EthR | | Authors: | Willand, N, Desroses, M, Toto, P, Diri, B, Lens, Z, Villeret, V, Rucktooa, P, Locht, C, Baulard, A, Deprez, B. | | Deposit date: | 2010-08-03 | | Release date: | 2010-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exploring drug target flexibility using in situ click chemistry: application to a mycobacterial transcriptional regulator.
Acs Chem.Biol., 5, 2010
|
|
3Q0U
 
 | | EthR from Mycobacterium tuberculosis in complex with compound BDM31379 | | Descriptor: | 2-phenyl-1-{4-[3-(thiophen-2-yl)-1,2,4-oxadiazol-5-yl]piperidin-1-yl}ethanone, HTH-type transcriptional regulator EthR | | Authors: | Flipo, M, Desroses, M, Dirie, B, Carette, X, Lens, Z, Rucktooa, P, Leroux, F, Piveteau, C, Demirkaya, F, Locht, C, Villeret, V, Christophe, T, Jeon, H.K, Brodin, P, Deprez, B, Baulard, A, Willand, N. | | Deposit date: | 2010-12-16 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design, synthesis and optimization of new EthR inhibitors. A new alternative approach to fight tuberculosis by boosting ethionamide
To be Published
|
|
3Q3S
 
 | |
1EI5
 
 | | CRYSTAL STRUCTURE OF A D-AMINOPEPTIDASE FROM OCHROBACTRUM ANTHROPI | | Descriptor: | D-AMINOPEPTIDASE | | Authors: | Bompard-Gilles, C, Remaut, H, Villeret, V, Prange, T, Fanuel, L, Joris, J, Frere, J.-M, Van Beeumen, J. | | Deposit date: | 2000-02-24 | | Release date: | 2000-10-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a D-aminopeptidase from Ochrobactrum anthropi, a new member of the 'penicillin-recognizing enzyme' family.
Structure Fold.Des., 8, 2000
|
|
7NTQ
 
 | | Crystal structure of the SARS-CoV-2 Main Protease complexed with N-(pyridin-3-ylmethyl)thioformamide | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Dupre, E, Villeret, V, Hanoulle, X. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.495 Å) | | Cite: | Novel dithiocarbamates selectively inhibit 3CL protease of SARS-CoV-2 and other coronaviruses.
Eur.J.Med.Chem., 250, 2023
|
|
7NTS
 
 | | Crystal structure of the SARS-CoV-2 Main Protease with oxidized C145 | | Descriptor: | DIMETHYL SULFOXIDE, FORMIC ACID, GLYCEROL, ... | | Authors: | Dupre, E, Villeret, V, Hanoulle, X. | | Deposit date: | 2021-03-10 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.477 Å) | | Cite: | NMR Spectroscopy of the Main Protease of SARS-CoV-2 and Fragment-Based Screening Identify Three Protein Hotspots and an Antiviral Fragment.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
