7PTV
 
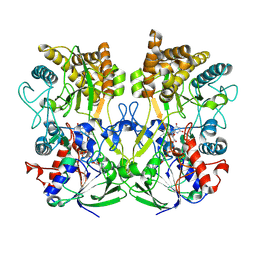 | | Structure of the Mimivirus genomic fibre asymmetric unit | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative glucose-methanol-choline oxidoreductase protein | | Authors: | Villalta, A, Schmitt, A, Estrozi, L.F, Quemin, E.R.J, Alempic, J.M, Lartigue, A, Prazak, V, Belmudes, L, Vasishtan, D, Colmant, A.M.G, Honore, F.A, Coute, Y, Grunewald, K, Abergel, C. | | Deposit date: | 2021-09-27 | | Release date: | 2022-08-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The giant mimivirus 1.2 Mb genome is elegantly organized into a 30 nm diameter helical protein shield.
Elife, 11, 2022
|
|
7YX5
 
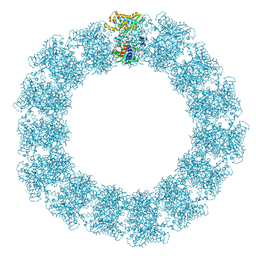 | | Structure of the Mimivirus genomic fibre in its relaxed 5-start helix form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative glucose-methanol-choline oxidoreductase protein | | Authors: | Villalta, A, Schmitt, A, Estrozi, L.F, Quemin, E.R.J, Alempic, J.M, Lartigue, A, Prazak, V, Belmudes, L, Vasishtan, D, Colmant, A.M.G, Honore, F.A, Coute, Y, Grunewald, K, Abergel, C. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The giant mimivirus 1.2 Mb genome is elegantly organized into a 30 nm diameter helical protein shield.
Elife, 11, 2022
|
|
7YX3
 
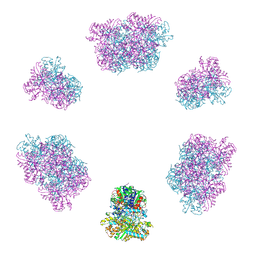 | | Structure of the Mimivirus genomic fibre in its compact 6-start helix form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative GMC-type oxidoreductase | | Authors: | Villalta, A, Schmitt, A, Estrozi, L.F, Quemin, E.R.J, Alempic, J.M, Lartigue, A, Prazak, V, Belmudes, L, Vasishtan, D, Colmant, A.M.G, Honore, F.A, Coute, Y, Grunewald, K, Abergel, C. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The giant mimivirus 1.2 Mb genome is elegantly organized into a 30-nm diameter helical protein shield.
Elife, 11, 2022
|
|
7YX4
 
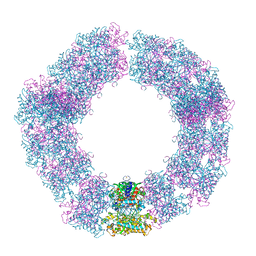 | | Structure of the Mimivirus genomic fibre in its compact 5-start helix form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative glucose-methanol-choline oxidoreductase protein | | Authors: | Villalta, A, Schmitt, A, Estrozi, L.F, Quemin, E.R.J, Alempic, J.M, Lartigue, A, Prazak, V, Belmudes, L, Vasishtan, D, Colmant, A.M.G, Honore, F.A, Coute, Y, Grunewald, K, Abergel, C. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The giant mimivirus 1.2 Mb genome is elegantly organized into a 30 nm diameter helical protein shield.
Elife, 11, 2022
|
|
1ZO8
 
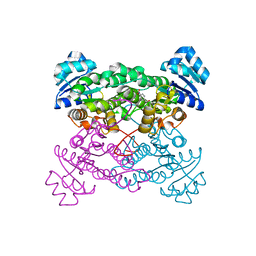 | | X-ray Structure of the haloalcohol dehalogenase HheC of Agrobacterium radiobacter AD1 in complex with (S)-para-nitrostyrene oxide, with a water molecule in the halide-binding site | | Descriptor: | (S)-PARA-NITROSTYRENE OXIDE, halohydrin dehalogenase | | Authors: | de Jong, R.M, Tiesinga, J.J.W, Tang, L, Villa, A, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2005-05-12 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for the Enantioselectivity of an Epoxide Ring Opening Reaction Catalyzed by Halo Alcohol Dehalogenase HheC.
J.Am.Chem.Soc., 127, 2005
|
|
8AS0
 
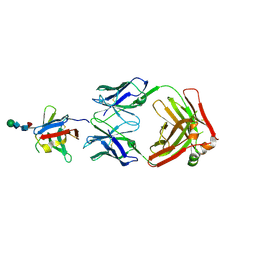 | | PD-1 extracellular domain in complex with Fab fragment from D12 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ongaro, T, Scietti, L, Pluss, L, Peissert, F, Villa, A, Puca, E, De Luca, R, Neri, D, Forneris, F. | | Deposit date: | 2022-08-17 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Selection of a PD-1 blocking antibody from a novel fully human phage display library.
Protein Sci., 31, 2022
|
|
7AH1
 
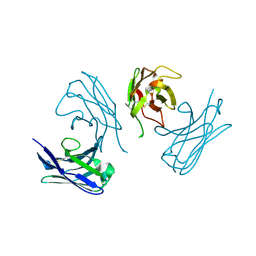 | | L19 diabody fragment from immunocytokine L19-IL2 | | Descriptor: | Anti-(ED-B) scFV | | Authors: | Ongaro, T, Guarino, S.R, Scietti, L, Palamini, M, Wulhfard, S, Villa, A, Neri, D, Forneris, F. | | Deposit date: | 2020-09-23 | | Release date: | 2021-02-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inference of molecular structure for characterization and improvement of clinical grade immunocytokines.
J.Struct.Biol., 213, 2021
|
|
8UPY
 
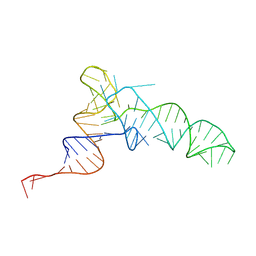 | | Methanosarcine mazei tRNAPyl in A-site of ribosome | | Descriptor: | RNA (72-MER) | | Authors: | Krahn, N, Zhang, J, Melnikov, S.V, Tharp, J.M, Villa, A, Patel, A, Howard, R.J, Gabir, H, Patel, T.R, Stetefeld, J, Puglisi, J, Soll, D. | | Deposit date: | 2023-10-23 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | tRNA shape is an identity element for an archaeal pyrrolysyl-tRNA synthetase from the human gut.
Nucleic Acids Res., 52, 2024
|
|
8UPT
 
 | | Candidatus Methanomethylophilus alvus tRNAPyl in A-site of ribosome | | Descriptor: | RNA (71-MER) | | Authors: | Krahn, N, Zhang, J, Melnikov, S.V, Tharp, J.M, Villa, A, Patel, A, Howard, R.J, Gabir, H, Patel, T.R, Stetefeld, J, Puglisi, J, Soll, D. | | Deposit date: | 2023-10-23 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | tRNA shape is an identity element for an archaeal pyrrolysyl-tRNA synthetase from the human gut.
Nucleic Acids Res., 52, 2024
|
|
1ZMT
 
 | | Structure of haloalcohol dehalogenase HheC of Agrobacterium radiobacter AD1 in complex with (R)-para-nitro styrene oxide, with a water molecule in the halide-binding site | | Descriptor: | (R)-PARA-NITROSTYRENE OXIDE, Haloalcohol dehalogenase HheC | | Authors: | de Jong, R.M, Tiesinga, J.J.W, Villa, A, Tang, L, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2005-05-10 | | Release date: | 2005-10-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for the Enantioselectivity of an Epoxide Ring Opening Reaction Catalyzed by Halo Alcohol Dehalogenase HheC.
J.Am.Chem.Soc., 127, 2005
|
|
1CA4
 
 | | STRUCTURE OF TNF RECEPTOR ASSOCIATED FACTOR 2 (TRAF2) | | Descriptor: | PROTEIN (TNF RECEPTOR ASSOCIATED FACTOR 2) | | Authors: | Park, Y.C, Burkitt, V, Villa, A.R, Tong, L, Wu, H. | | Deposit date: | 1999-02-23 | | Release date: | 1999-04-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for self-association and receptor recognition of human TRAF2.
Nature, 398, 1999
|
|
1CA9
 
 | | STRUCTURE OF TNF RECEPTOR ASSOCIATED FACTOR 2 IN COMPLEX WITH A PEPTIDE FROM TNF-R2 | | Descriptor: | PROTEIN (TNF RECEPTOR ASSOCIATED FACTOR 2), PROTEIN (TNF-R2) | | Authors: | Park, Y.C, Burkitt, V, Villa, A.R, Tong, L, Wu, H. | | Deposit date: | 1999-02-25 | | Release date: | 1999-04-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for self-association and receptor recognition of human TRAF2.
Nature, 398, 1999
|
|
5VLO
 
 | | The structure of human CamKII with bound inhibitor | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Calcium/calmodulin-dependent protein kinase type II subunit delta, N-[(2S)-2-(diethylamino)propyl]-2-[(2S)-2-(methylcarbamoyl)azetidin-1-yl]-6-[5-(thiophen-2-yl)pyrazolo[1,5-a]pyrimidin-3-yl]pyridine-4-carboxamide | | Authors: | Somoza, J.R, Villasenor, A.G. | | Deposit date: | 2017-04-25 | | Release date: | 2017-08-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Inhibitors of CamKII
To Be Published
|
|
6DGT
 
 | | Selective PI3K beta inhibitor bound to PI3K delta | | Descriptor: | 4-[1-(5,8-difluoroquinolin-4-yl)-2-methyl-4-(4H-1,2,4-triazol-3-yl)-1H-benzimidazol-6-yl]-3-fluoropyridin-2-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Somoza, J, Villasenor, A, McGrath, M. | | Deposit date: | 2018-05-18 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Atropisomerism by Design: Discovery of a Selective and Stable Phosphoinositide 3-Kinase (PI3K) beta Inhibitor.
J. Med. Chem., 61, 2018
|
|
1LOX
 
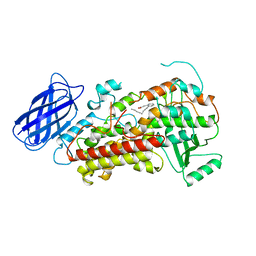 | | RABBIT RETICULOCYTE 15-LIPOXYGENASE | | Descriptor: | (2E)-3-(2-OCT-1-YN-1-YLPHENYL)ACRYLIC ACID, 15-LIPOXYGENASE, FE (II) ION | | Authors: | Gillmor, S.A, Villasenor, A, Fletterick, R.J, Sigal, E, Browner, M.F. | | Deposit date: | 1997-10-06 | | Release date: | 1998-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of mammalian 15-lipoxygenase reveals similarity to the lipases and the determinants of substrate specificity.
Nat.Struct.Biol., 4, 1997
|
|
4MIB
 
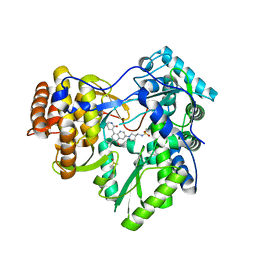 | | Hepatitis C Virus polymerase NS5B genotype 1b (BK) in complex with Compound 48 (N-({(3S)-1-[6-tert-butyl-5-methoxy-8-(2-oxo-1,2-dihydropyridin-3-yl)quinolin-3-yl]pyrrolidin-3-yl}methyl)methanesulfonamide) | | Descriptor: | DIMETHYL SULFOXIDE, N-({(3S)-1-[6-tert-butyl-5-methoxy-8-(2-oxo-1,2-dihydropyridin-3-yl)quinolin-3-yl]pyrrolidin-3-yl}methyl)methanesulfonamide, RNA-DIRECTED RNA POLYMERASE | | Authors: | Harris, S.F, Villasenor, A.G. | | Deposit date: | 2013-08-30 | | Release date: | 2014-05-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of N-[4-[6-tert-butyl-5-methoxy-8-(6-methoxy-2-oxo-1H-pyridin-3-yl)-3-quinolyl]phenyl]methanesulfonamide (RG7109), a potent inhibitor of the hepatitis C virus NS5B polymerase.
J.Med.Chem., 57, 2014
|
|
4UNK
 
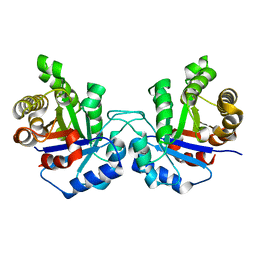 | | Crystal structure of human triosephosphate isomerase (mutant N15D) | | Descriptor: | TRIOSEPHOSPHATE ISOMERASE | | Authors: | DeLaMora-DeLaMora, I, Torres-Larios, A, Enriquez-Flores, S, Mendez, S.T, Castillo-Villanueva, A, Gomez-Manzo, S, Lopez-Velazquez, G, Marcial-Quino, J, Torres-Arroyo, A, Garcia-Torres, I, Reyes-Vivas, H, Oria-Hernandez, J. | | Deposit date: | 2014-05-29 | | Release date: | 2015-02-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Human Triosephosphate Isomerase (Mutant N15D)
To be Published
|
|
4UNL
 
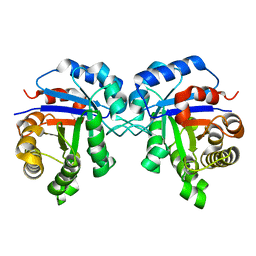 | | Crystal structure of a single mutant (N71D) of triosephosphate isomerase from human | | Descriptor: | TRIOSEPHOSPHATE ISOMERASE | | Authors: | DeLaMora-DeLaMora, I, Torres-Larios, A, Enriquez-Flores, S, Mendez, S.T, Castillo-Villanueva, A, Gomez-Manzo, S, Lopez-Velazquez, G, Marcial-Quino, J, Torres-Arroyo, A, Garcia-Torres, I, Reyes-Vivas, H, Oria-Hernandez, J. | | Deposit date: | 2014-05-29 | | Release date: | 2015-02-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of a Single Mutant (N71D) of Triosephosphate Isomerase from Human
To be Published
|
|
5NCR
 
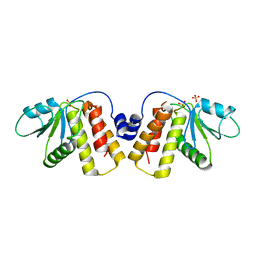 | | OH1 from the Orf virus: a tyrosine phosphatase that displays distinct structural features and triple substrate specificity | | Descriptor: | PHOSPHATE ION, SULFATE ION, tyrosine phosphatase | | Authors: | Segovia, D, Haouz, A, Berois, M, Villarino, A, Andre-Leroux, G. | | Deposit date: | 2017-03-06 | | Release date: | 2017-08-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | OH1 from Orf Virus: A New Tyrosine Phosphatase that Displays Distinct Structural Features and Triple Substrate Specificity.
J. Mol. Biol., 429, 2017
|
|
3FFI
 
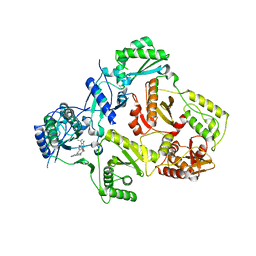 | | HIV-1 RT with pyridone non-nucleoside inhibitor | | Descriptor: | 3-chloro-5-({6-[2-(3,4-dihydroisoquinolin-2(1H)-yl)-2-oxoethyl]-3-(dimethylamino)-2-oxo-1,2-dihydropyridin-4-yl}oxy)benzonitrile, RT p51, Reverse transcriptase/ribonuclease H | | Authors: | Harris, S.F, Villasenor, A. | | Deposit date: | 2008-12-03 | | Release date: | 2009-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Pyridone Diaryl Ether Non-Nucleoside Inhibitors of HIV-1 Reverse Transcriptase
To be Published
|
|
5I4U
 
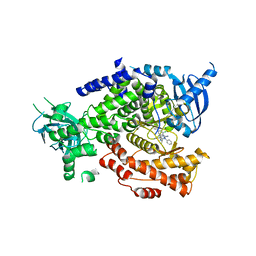 | | The crystal structure of PI3Kdelta with compound 34 | | Descriptor: | 2,4-diamino-6-{[(1S)-1-(5-chloro-4-oxo-3-phenyl-3,4-dihydroquinazolin-2-yl)ethyl]amino}pyrimidine-5-carbonitrile, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Somoza, J.R, Villasenor, A.G. | | Deposit date: | 2016-02-12 | | Release date: | 2017-02-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.372 Å) | | Cite: | The Design and Synthesis of Potent, Selective and Metabolically Stable PI3K[delta] Inhibitors
To be published
|
|
4BR1
 
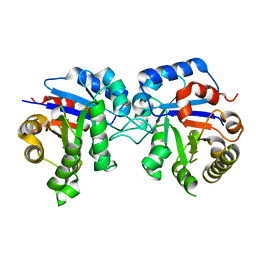 | | Protease-induced heterodimer of human triosephosphate isomerase. | | Descriptor: | TRIOSEPHOSPHATE ISOMERASE | | Authors: | DeLaMora-DeLaMora, I, Torres-Larios, A, Hernandez-Alcantara, G, Mendoza-Hernandez, G, Enriquez-Flores, S, Mendez, S.T, Castillo-Villanueva, A, Garcia-Torres, I, Torres-Arroyo, A, Gomez-Manzo, S, Marcial-Quino, J, Oria-Hernandez, J, Lopez-Velazquez, G, Reyes-Vivas, H. | | Deposit date: | 2013-06-03 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Differential Proteolysis in Human Triosephosphate Isomerase
To be Published
|
|
6WML
 
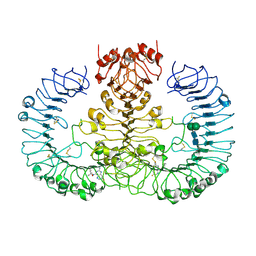 | | Human TLR8 bound to the potent agonist, GS-9688 (Selgantolimod) | | Descriptor: | (2R)-2-[(2-amino-7-fluoropyrido[3,2-d]pyrimidin-4-yl)amino]-2-methylhexan-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Toll-like receptor 8, ... | | Authors: | Appleby, T.C, Perry, J.K, Mish, M, Villasenor, A.G, Mackman, R.L. | | Deposit date: | 2020-04-21 | | Release date: | 2021-02-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of GS-9688 (Selgantolimod) as a Potent and Selective Oral Toll-Like Receptor 8 Agonist for the Treatment of Chronic Hepatitis B.
J.Med.Chem., 63, 2020
|
|
5DFW
 
 | |
7PEG
 
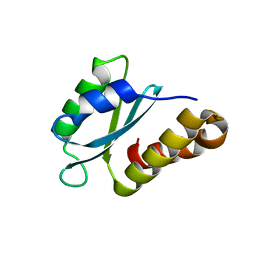 | | Structure of the sporulation/germination protein YhcN from Bacillus subtilis | | Descriptor: | Probable spore germination lipoprotein YhcN | | Authors: | Liu, B, Chan, H, Bauda, E, Contreras-Martel, C, Bellard, L, Villard, A.M, Mas, C, Neumann, E, Fenel, D, Favier, A, Serrano, M, Henriques, A.O.H, Rodrigues, C.D.A, Morlot, C. | | Deposit date: | 2021-08-10 | | Release date: | 2022-08-24 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural insights into ring-building motif domains involved in bacterial sporulation.
J.Struct.Biol., 214, 2022
|
|
