3HAV
 
 | | Structure of the streptomycin-ATP-APH(2")-IIa ternary complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Aminoglycoside phosphotransferase, MAGNESIUM ION, ... | | Authors: | Young, P.G, Baker, E.N, Vakulenko, S.B, Smith, C.A. | | Deposit date: | 2009-05-02 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The crystal structures of substrate and nucleotide complexes of Enterococcus faecium aminoglycoside-2''-phosphotransferase-IIa [APH(2'')-IIa] provide insights into substrate selectivity in the APH(2'') subfamily.
J.Bacteriol., 191, 2009
|
|
4ZDX
 
 | | Structure of OXA-51 beta-lactamase | | Descriptor: | 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, Beta-lactamase, GLYCEROL | | Authors: | Smith, C.A, Antunes, N.T, Stewart, N.K, Frase, H, Toth, M, Kantardjieff, K.A, Vakulenko, S.B. | | Deposit date: | 2015-04-20 | | Release date: | 2015-06-17 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural Basis for Enhancement of Carbapenemase Activity in the OXA-51 Family of Class D beta-Lactamases.
Acs Chem.Biol., 10, 2015
|
|
5IY2
 
 | | Structure of apo OXA-143 carbapenemase | | Descriptor: | Beta-lactamase OXA-143, GLYCEROL | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2016-03-23 | | Release date: | 2017-08-09 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | The role of conserved surface hydrophobic residues in the carbapenemase activity of the class D beta-lactamases.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
8FAJ
 
 | | OXA-48-NA-1-157 inhibitor complex | | Descriptor: | (5R)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-5-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase, CADMIUM ION, ... | | Authors: | Smith, C.A, Stewart, N.K, Toth, M, Vakulenko, S.B. | | Deposit date: | 2022-11-28 | | Release date: | 2023-10-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The C5 alpha-Methyl-Substituted Carbapenem NA-1-157 Exhibits Potent Activity against Klebsiella spp. Isolates Producing OXA-48-Type Carbapenemases.
Acs Infect Dis., 9, 2023
|
|
8DA2
 
 | | Acinetobacter baumannii L,D-transpeptidase | | Descriptor: | L,D-transpeptidase family protein | | Authors: | Toth, M, Stewart, N.K, Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2022-06-12 | | Release date: | 2022-09-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The l,d-Transpeptidase Ldt Ab from Acinetobacter baumannii Is Poorly Inhibited by Carbapenems and Has a Unique Structural Architecture.
Acs Infect Dis., 8, 2022
|
|
3HAM
 
 | | Structure of the gentamicin-APH(2")-IIa complex | | Descriptor: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, Aminoglycoside phosphotransferase, GLYCEROL | | Authors: | Young, P.G, Baker, E.N, Vakulenko, S.B, Smith, C.A. | | Deposit date: | 2009-05-02 | | Release date: | 2009-07-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structures of substrate and nucleotide complexes of Enterococcus faecium aminoglycoside-2''-phosphotransferase-IIa [APH(2'')-IIa] provide insights into substrate selectivity in the APH(2'') subfamily.
J.Bacteriol., 191, 2009
|
|
1EWZ
 
 | | CRYSTAL STRUCTURE OF THE OXA-10 BETA-LACTAMASE FROM PSEUDOMONAS AERUGINOSA | | Descriptor: | BETA LACTAMASE OXA-10 | | Authors: | Golemi, D, Maveyraud, L, Vakulenko, S, Tranier, S, Ishiwata, A, Kotra, L.P, Samama, J.P, Mobashery, S. | | Deposit date: | 2000-04-28 | | Release date: | 2000-11-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The First Structural and Mechanistic Insights for Class D beta-Lactamases: Evidence for a Novel Catalytic Process for Turnover of beta-Lactam Antibiotics
J.Am.Chem.Soc., 122, 2000
|
|
1YT4
 
 | | Crystal structure of TEM-76 beta-lactamase at 1.4 Angstrom resolution | | Descriptor: | Beta-lactamase TEM | | Authors: | Thomas, V.L, Golemi-Kotra, D, Kim, C, Vakulenko, S.B, Mobashery, S, Shoichet, B.K. | | Deposit date: | 2005-02-09 | | Release date: | 2005-07-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Consequences of the Inhibitor-Resistant Ser130Gly Substitution in TEM beta-Lactamase.
Biochemistry, 44, 2005
|
|
6CTZ
 
 | | Structure of the GDP and kanamycin complex of APH(2")-IIia | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, Gentamicin resistance protein, ... | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2018-03-23 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structural basis for the diversity of the mechanism of nucleotide hydrolysis by the aminoglycoside-2''-phosphotransferases
Acta Crystallogr.,Sect.D, 75, 2019
|
|
6EDM
 
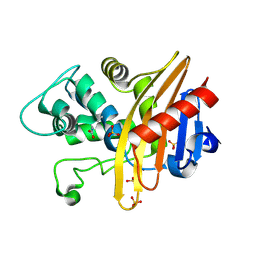 | | Structure of apo-CDD-1 beta-lactamase | | Descriptor: | Beta-lactamase, SULFATE ION | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2018-08-09 | | Release date: | 2019-08-14 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The crystal structures of CDD-1, the intrinsic class D beta-lactamase from the pathogenic Gram-positive bacterium Clostridioides difficile, and its complex with cefotaxime.
J.Struct.Biol., 208, 2019
|
|
9MTV
 
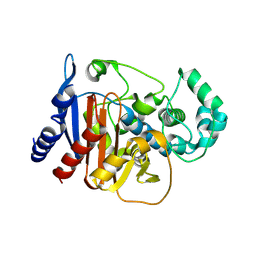 | |
9MTU
 
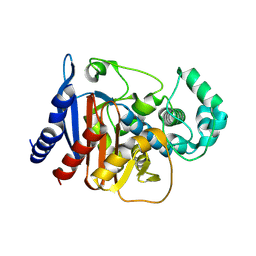 | |
8V9G
 
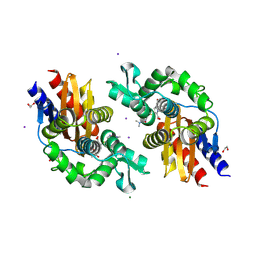 | | GES-5-meropenem complex | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, 1,2-ETHANEDIOL, IODIDE ION, ... | | Authors: | Smith, C.A, Stewart, N.K, Vakulenko, S.B. | | Deposit date: | 2023-12-08 | | Release date: | 2024-04-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Restricted Rotational Flexibility of the C5 alpha-Methyl-Substituted Carbapenem NA-1-157 Leads to Potent Inhibition of the GES-5 Carbapenemase.
Acs Infect Dis., 10, 2024
|
|
2QPN
 
 | | GES-1 beta-lactamase | | Descriptor: | Beta-lactamase GES-1, SULFATE ION | | Authors: | Smith, C.A, Caccamo, M, Kantardjieff, K.A, Vakulenko, S. | | Deposit date: | 2007-07-24 | | Release date: | 2008-08-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure of GES-1 at atomic resolution: insights into the evolution of carbapenamase activity in the class A extended-spectrum beta-lactamases.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
6W5F
 
 | | Class D beta-lactamase BSU-2 delta mutant | | Descriptor: | 1,2-ETHANEDIOL, BSU-2delta mutant, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Smith, C.A, Vakulenko, S.B, Stewart, N.K, Toth, M. | | Deposit date: | 2020-03-13 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A surface loop modulates activity of the Bacillus class D beta-lactamases.
J.Struct.Biol., 211, 2020
|
|
6W5O
 
 | | Class D beta-lactamase BAT-2 delta mutant | | Descriptor: | 1,2-ETHANEDIOL, BAT-2 Beta-lactamase delta mutant, CITRATE ANION | | Authors: | Smith, C.A, Vakulenko, S.B, Stewart, N.K, Toth, M. | | Deposit date: | 2020-03-13 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A surface loop modulates activity of the Bacillus class D beta-lactamases.
J.Struct.Biol., 211, 2020
|
|
6W5G
 
 | | Class D beta-lactamase BAT-2 | | Descriptor: | 1,2-ETHANEDIOL, BAT-2 beta-lactamase | | Authors: | Smith, C.A, Vakulenko, S.B, Stewart, N.K, Toth, M. | | Deposit date: | 2020-03-13 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | A surface loop modulates activity of the Bacillus class D beta-lactamases.
J.Struct.Biol., 211, 2020
|
|
6W5E
 
 | | Class D beta-lactamase BSU-2 | | Descriptor: | 1,2-ETHANEDIOL, BSU-2 beta-lactamase, MALONATE ION | | Authors: | Smith, C.A, Vakulenko, S.B, Stewart, N.K, Toth, M. | | Deposit date: | 2020-03-13 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A surface loop modulates activity of the Bacillus class D beta-lactamases.
J.Struct.Biol., 211, 2020
|
|
3TDV
 
 | |
9MTW
 
 | | Crystal structure of ADC-1-ertapenem complex | | Descriptor: | (2S,3R,4R)-4-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, (4R,5S)-3-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, 1,2-ETHANEDIOL, ... | | Authors: | Smith, C.A, Stewart, N.K, Vakulenko, S.B. | | Deposit date: | 2025-01-12 | | Release date: | 2025-04-23 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Evolution of carbapenemase activity in the class C beta-lactamase ADC-1.
Mbio, 2025
|
|
3TDW
 
 | |
6BFF
 
 | |
6BFH
 
 | |
8V9H
 
 | | GES-5-NA-1-157 complex | | Descriptor: | (5R)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-5-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Smith, C.A, Stewart, N.K, Vakulenko, S.B. | | Deposit date: | 2023-12-08 | | Release date: | 2024-04-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Restricted Rotational Flexibility of the C5 alpha-Methyl-Substituted Carbapenem NA-1-157 Leads to Potent Inhibition of the GES-5 Carbapenemase.
Acs Infect Dis., 10, 2024
|
|
5CTM
 
 | | Structure of BPu1 beta-lactamase | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CITRATE ANION, ... | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2015-07-24 | | Release date: | 2015-11-18 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Class D beta-lactamases do exist in Gram-positive bacteria.
Nat.Chem.Biol., 12, 2016
|
|
