8J3R
 
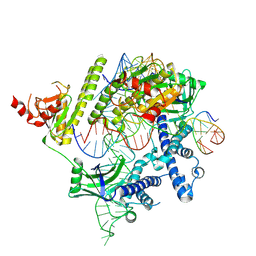 | | Cryo-EM structure of the AsCas12f-HKRA-sgRNAS3-5v7-target DNA | | Descriptor: | DNA (37-MER), DNA (38-MER), MAGNESIUM ION, ... | | Authors: | Hino, T, Omura, N.S, Nakagawa, R, Togashi, T, Takeda, N.S, Hiramoto, T, Tasaka, S, Hirano, H, Tokuyama, T, Uosaki, H, Ishiguro, H, Yamano, H, Ozaki, Y, Motooka, D, Mori, H, Kirita, Y, Kise, Y, Itoh, Y, Matoba, S, Aburatani, H, Yachie, N, Siksnys, V, Ohmori, T, Hoshino, A, Nureki, O. | | Deposit date: | 2023-04-18 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | An AsCas12f-based compact genome-editing tool derived by deep mutational scanning and structural analysis.
Cell, 186, 2023
|
|
1D8K
 
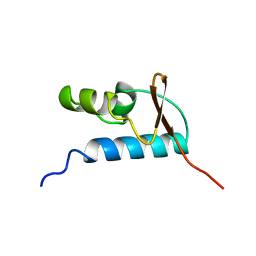 | | SOLUTION STRUCTURE OF THE CENTRAL CORE DOMAIN OF TFIIE BETA | | Descriptor: | GENERAL TRANSCRIPTION FACTOR TFIIE-BETA | | Authors: | Okuda, M, Watanabe, Y, Okamura, H, Hanaoka, F, Ohkuma, Y, Nishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-10-25 | | Release date: | 2000-04-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the central core domain of TFIIEbeta with a novel double-stranded DNA-binding surface.
EMBO J., 19, 2000
|
|
1D8J
 
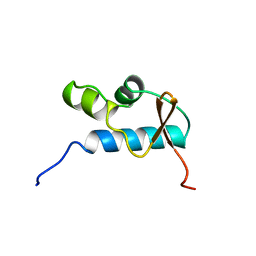 | | SOLUTION STRUCTURE OF THE CENTRAL CORE DOMAIN OF TFIIE BETA | | Descriptor: | GENERAL TRANSCRIPTION FACTOR TFIIE-BETA | | Authors: | Okuda, M, Watanabe, Y, Okamura, H, Hanaoka, F, Ohkuma, Y, Nishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-10-25 | | Release date: | 2000-04-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the central core domain of TFIIEbeta with a novel double-stranded DNA-binding surface.
EMBO J., 19, 2000
|
|
8UH8
 
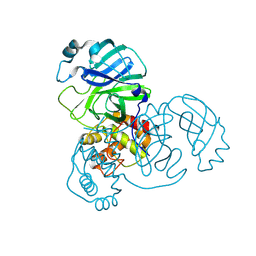 | | Crystal structure of SARS-CoV-2 main protease E166V (Apo structure) | | Descriptor: | ORF1a polyprotein | | Authors: | Bulut, H, Hayashi, H, Kuwata, N, Tsuji, K, Das, D, Tamamura, H, Mitsuya, H. | | Deposit date: | 2023-10-07 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | TKB272, an Orally Available SARS-CoV-2-Mpro Inhibitor Containing 5-Fluorobenzothiazole, Potently Blocks SARS-CoV-2 Replication without Ritonavir
To Be Published
|
|
8UH5
 
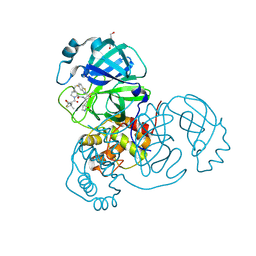 | | Crystal structure of SARS-CoV-2 main protease in complex with an inhibitor TKB-272 | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(5-fluoro-1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER | | Authors: | Bulut, H, Hayashi, H, Kuwata, N, Tsuji, K, Das, D, Tamamura, H, Mitsuya, H. | | Deposit date: | 2023-10-06 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | TKB272, an Orally Available SARS-CoV-2-Mpro Inhibitor Containing 5-Fluorobenzothiazole, Potently Blocks SARS-CoV-2 Replication without Ritonavir
To Be Published
|
|
8UH9
 
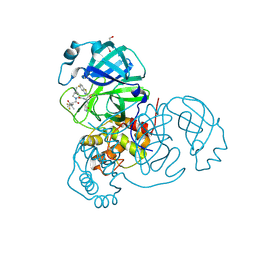 | | Crystal structure of SARS-CoV-2 main protease E166V mutant in complex with an inhibitor TKB-272 | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(5-fluoro-1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER | | Authors: | Bulut, H, Hayashi, H, Kuwata, N, Tsuji, K, Das, D, Tamamura, H, Mitsuya, H. | | Deposit date: | 2023-10-07 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.067 Å) | | Cite: | TKB272, an Orally Available SARS-CoV-2-Mpro Inhibitor Containing 5-Fluorobenzothiazole, Potently Blocks SARS-CoV-2 Replication without Ritonavir
To Be Published
|
|
1VTU
 
 | | Structural characteristics of enantiomorphic DNA: Crystal analysis of racemates of the D(CGCGCG) duplex | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*CP*G)-3') | | Authors: | Doi, M, Inoue, M, Tomoo, K, Ishida, T, Ueda, Y, Akagi, M, Urata, H. | | Deposit date: | 1994-08-29 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Characteristics of Enantiomorphic DNA: Crystal Analysis of Racemates of the d(CGCGCG) Duplex
J.Am.Chem.Soc., 115, 1993
|
|
5Y5M
 
 | | SFX structure of cytochrome P450nor: a complete dark data without pump laser (resting state) | | Descriptor: | NADP nitrous oxide-forming nitric oxide reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tosha, T, Nomura, T, Nishida, T, Saeki, N, Okubayashi, K, Yamagiwa, R, Sugahara, M, Nakane, T, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Mizohata, E, Yamashita, A, Kanematsu, Y, Takano, Y, Nango, E, Tanaka, R, Nureki, O, Ikemoto, Y, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Iwata, S, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
5Y5L
 
 | | Time-resolved SFX structure of cytochrome P450nor: dark-2 data in the absence of NADH (resting state) | | Descriptor: | NADP nitrous oxide-forming nitric oxide reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tosha, T, Nomura, T, Nishida, T, Saeki, N, Okubayashi, K, Yamagiwa, R, Sugahara, M, Nakane, T, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Mizohata, E, Yamashita, A, Kanematsu, Y, Takano, Y, Nango, E, Tanaka, R, Nureki, O, Ikemoto, Y, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Iwata, S, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
5Y5H
 
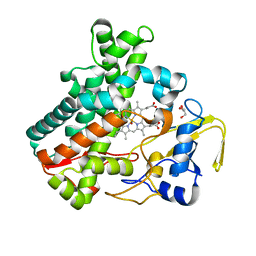 | | SF-ROX structure of cytochrome P450nor (NO-bound state) determined at SACLA | | Descriptor: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, NITRIC OXIDE, ... | | Authors: | Tosha, T, Nomura, T, Nishida, T, Yamagiwa, R, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Yamashita, A, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
5Y5I
 
 | | Time-resolved SFX structure of cytochrome P450nor: 20 ms after photo-irradiation of caged NO in the presence of NADH (NO-bound state), light data | | Descriptor: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, NITRIC OXIDE, ... | | Authors: | Tosha, T, Nomura, T, Nishida, T, Saeki, N, Okubayashi, K, Yamagiwa, R, Sugahara, M, Nakane, T, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Mizohata, E, Yamashita, A, Kanematsu, Y, Takano, Y, Nango, E, Tanaka, R, Nureki, O, Ikemoto, Y, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Iwata, S, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
5Y5J
 
 | | Time-resolved SFX structure of cytochrome P450nor: dark-2 data in the presence of NADH (resting state) | | Descriptor: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tosha, T, Nomura, T, Nishida, T, Saeki, N, Okubayashi, K, Yamagiwa, R, Sugahara, M, Nakane, T, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Mizohata, E, Yamashita, A, Kanematsu, Y, Takano, Y, Nango, E, Tanaka, R, Nureki, O, Ikemoto, Y, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Iwata, S, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
5Y5K
 
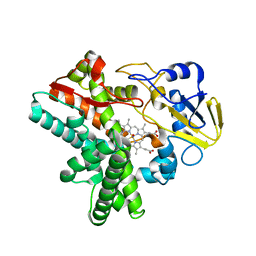 | | Time-resolved SFX structure of cytochrome P450nor : 20 ms after photo-irradiation of caged NO in the absence of NADH (NO-bound state), light data | | Descriptor: | NADP nitrous oxide-forming nitric oxide reductase, NITRIC OXIDE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tosha, T, Nomura, T, Nishida, T, Saeki, N, Okubayashi, K, Yamagiwa, R, Sugahara, M, Nakane, T, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Mizohata, E, Yamashita, A, Kanematsu, Y, Takano, Y, Nango, E, Tanaka, R, Nureki, O, Ikemoto, Y, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Iwata, S, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
5X1B
 
 | | CO bound cytochrome c oxidase at 20 nsec after pump laser irradiation to release CO from O2 reduction center | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Kubo, M, Baba, S, Yamashita, K, Hirata, K, Ueno, G, Nomura, T, Kimura, T, Shinzawa-Itoh, K, Baba, J, Hatano, K, Eto, Y, Miyamoto, A, Murakami, H, Kumasaka, T, Owada, S, Tono, K, Yabashi, M, Yamaguchi, Y, Yanagisawa, S, Sakaguchi, M, Ogura, T, Komiya, R, Yan, J, Yamashita, E, Yamamoto, M, Ago, H, Yoshikawa, S, Tsukihara, T. | | Deposit date: | 2017-01-25 | | Release date: | 2017-08-09 | | Last modified: | 2017-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A nanosecond time-resolved XFEL analysis of structural changes associated with CO release from cytochrome c oxidase.
Sci Adv, 3, 2017
|
|
5X19
 
 | | CO bound cytochrome c oxidase at 100 micro sec after pump laser irradiation to release CO from O2 reduction center | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Kubo, M, Baba, S, Yamashita, K, Hirata, K, Ueno, G, Nomura, T, Kimura, T, Shinzawa-Itoh, K, Baba, J, Hatano, K, Eto, Y, Miyamoto, A, Murakami, H, Kumasaka, T, Owada, S, Tono, K, Yabashi, M, Yamaguchi, Y, Yanagisawa, S, Sakaguchi, M, Ogura, T, Komiya, R, Yan, J, Yamashita, E, Yamamoto, M, Ago, H, Yoshikawa, S, Tsukihara, T. | | Deposit date: | 2017-01-25 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A nanosecond time-resolved XFEL analysis of structural changes associated with CO release from cytochrome c oxidase.
Sci Adv, 3, 2017
|
|
5X1F
 
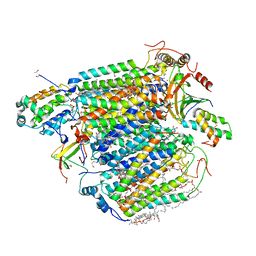 | | CO bound cytochrome c oxidase without pump laser irradiation at 278K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Kubo, M, Baba, S, Yamashita, K, Hirata, K, Ueno, G, Nomura, T, Kimura, T, Shinzawa-Itoh, K, Baba, J, Hatano, K, Eto, Y, Miyamoto, A, Murakami, H, Kumasaka, T, Owada, S, Tono, K, Yabashi, M, Yamaguchi, Y, Yanagisawa, S, Sakaguchi, M, Ogura, T, Komiya, R, Yan, J, Yamashita, E, Yamamoto, M, Ago, H, Yoshikawa, S, Tsukihara, T. | | Deposit date: | 2017-01-25 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A nanosecond time-resolved XFEL analysis of structural changes associated with CO release from cytochrome c oxidase.
Sci Adv, 3, 2017
|
|
1D06
 
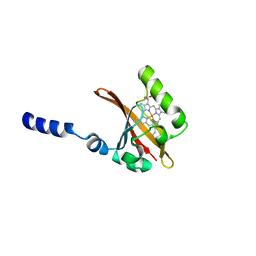 | | STRUCTURAL BASIS OF DIMERIZATION AND SENSORY MECHANISMS OF OXYGEN-SENSING DOMAIN OF RHIZOBIUM MELILOTI FIXL DETERMINED AT 1.4A RESOLUTION | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, nitrogen fixation regulatory protein fixL | | Authors: | Miyatake, H, Mukai, M, Park, S.-Y, Adachi, S, Tamura, K, Nakamura, H, Nakamura, K, Tsuchiya, T, Iizuka, T, Shiro, Y. | | Deposit date: | 1999-09-09 | | Release date: | 2000-03-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Sensory mechanism of oxygen sensor FixL from Rhizobium meliloti: crystallographic, mutagenesis and resonance Raman spectroscopic studies
J.MOL.BIOL., 301, 2000
|
|
1UAN
 
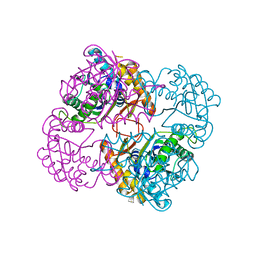 | | Crystal structure of the conserved protein TT1542 from Thermus thermophilus HB8 | | Descriptor: | hypothetical protein TT1542 | | Authors: | Handa, N, Terada, T, Tame, J.R.H, Park, S.-Y, Kinoshita, K, Ota, M, Nakamura, H, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-03-12 | | Release date: | 2003-08-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the conserved protein TT1542 from Thermus thermophilus HB8
PROTEIN SCI., 12, 2003
|
|
6KVQ
 
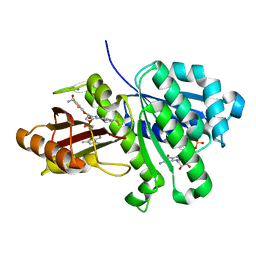 | | S. aureus FtsZ in complex with BOFP (compound 3) | | Descriptor: | CALCIUM ION, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Ferrer-Gonzalez, E, Fujita, J, Yoshizawa, T, Nelson, J.M, Pilch, A.J, Hillman, E, Ozawa, M, Kuroda, N, Parhi, A.K, LaVoie, E.J, Matsumura, H, Pilch, D.S. | | Deposit date: | 2019-09-05 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Design of a Fluorescent Probe for the Visualization of FtsZ in Clinically Important Gram-Positive and Gram-Negative Bacterial Pathogens.
Sci Rep, 9, 2019
|
|
6KVP
 
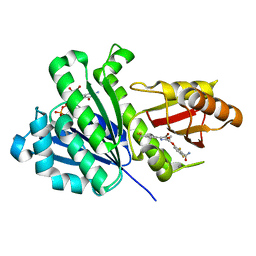 | | S. aureus FtsZ in complex with 3-(1-(5-bromo-4-(4-(trifluoromethyl)phenyl)oxazol-2-yl)ethoxy)-2,6-difluorobenzamide (compound 2) | | Descriptor: | 3-[(1R)-1-[5-bromanyl-4-[4-(trifluoromethyl)phenyl]-1,3-oxazol-2-yl]ethoxy]-2,6-bis(fluoranyl)benzamide, CALCIUM ION, Cell division protein FtsZ, ... | | Authors: | Ferrer-Gonzalez, E, Fujita, J, Yoshizawa, T, Nelson, J.M, Pilch, A.J, Hillman, E, Ozawa, M, Kuroda, N, Parhi, A.K, LaVoie, E.J, Matsumura, H, Pilch, D.S. | | Deposit date: | 2019-09-05 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Guided Design of a Fluorescent Probe for the Visualization of FtsZ in Clinically Important Gram-Positive and Gram-Negative Bacterial Pathogens.
Sci Rep, 9, 2019
|
|
7QY4
 
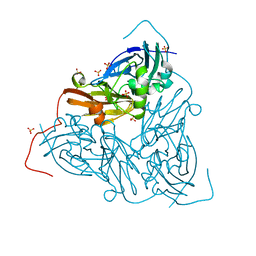 | | As isolated MSOX movie series dataset 5 (2 MGy) of the copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | Authors: | Rose, S.L, Baba, S, Okumura, H, Antonyuk, S.V, Sasaki, D, Tosha, T, Kumasaka, T, Eady, R.R, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2022-01-27 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Single crystal spectroscopy and multiple structures from one crystal (MSOX) define catalysis in copper nitrite reductases.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7QXK
 
 | | As isolated MSOX movie series dataset 1 (0.4 MGy) of the copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | Authors: | Rose, S.L, Baba, S, Okumura, H, Antonyuk, S.V, Sasaki, D, Tosha, T, Kumasaka, T, Eady, R.R, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2022-01-26 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Single crystal spectroscopy and multiple structures from one crystal (MSOX) define catalysis in copper nitrite reductases.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7QYC
 
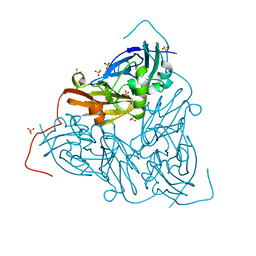 | | As isolated MSOX movie series dataset 20 (8 MGy) of the copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | Authors: | Rose, S.L, Baba, S, Okumura, H, Antonyuk, S.V, Sasaki, D, Tosha, T, Kumasaka, T, Eady, R.R, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2022-01-27 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Single crystal spectroscopy and multiple structures from one crystal (MSOX) define catalysis in copper nitrite reductases.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4UB8
 
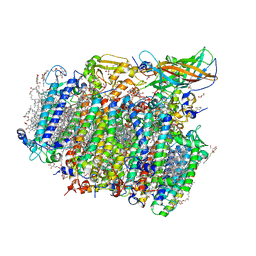 | | Native structure of photosystem II (dataset-2) by a femtosecond X-ray laser | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Akita, F, Hirata, K, Ueno, G, Murakami, H, Nakajima, Y, Shimizu, T, Yamashita, K, Yamamoto, M, Ago, H, Shen, J.R. | | Deposit date: | 2014-08-12 | | Release date: | 2014-12-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Native structure of photosystem II at 1.95 angstrom resolution viewed by femtosecond X-ray pulses.
Nature, 517, 2015
|
|
4UB6
 
 | | Native structure of photosystem II (dataset-1) by a femtosecond X-ray laser | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Akita, F, Hirata, K, Ueno, G, Murakami, H, Nakajima, Y, Shimizu, T, Yamashita, K, Yamamoto, M, Ago, H, Shen, J.R. | | Deposit date: | 2014-08-12 | | Release date: | 2014-12-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Native structure of photosystem II at 1.95 angstrom resolution viewed by femtosecond X-ray pulses.
Nature, 517, 2015
|
|
