7MOA
 
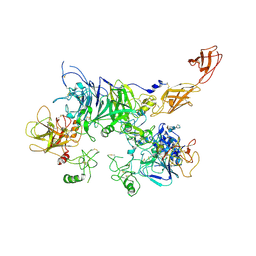 | | Cryo-EM structure of the c-MET II/HGF I complex bound with HGF II in a rigid conformation | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Hepatocyte growth factor, Hepatocyte growth factor receptor | | Authors: | Uchikawa, E, Chen, Z.M, Xiao, G.Y, Zhang, X.W, Bai, X.C. | | Deposit date: | 2021-05-01 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structural basis of the activation of c-MET receptor.
Nat Commun, 12, 2021
|
|
7MO7
 
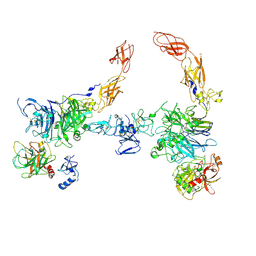 | | Cryo-EM structure of 2:2 c-MET/HGF holo-complex | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Hepatocyte growth factor, Hepatocyte growth factor receptor | | Authors: | Uchikawa, E, Chen, Z.M, Xiao, G.Y, Zhang, X.W, Bai, X.C. | | Deposit date: | 2021-05-01 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural basis of the activation of c-MET receptor.
Nat Commun, 12, 2021
|
|
7MO9
 
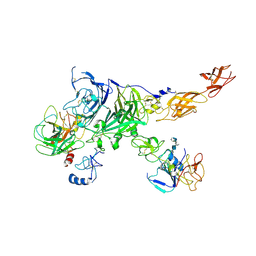 | | Cryo-EM map of the c-MET II/HGF I/HGF II (K4 and SPH) sub-complex | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Hepatocyte growth factor, Hepatocyte growth factor receptor | | Authors: | Uchikawa, E, Chen, Z.M, Xiao, G.Y, Zhang, X.W, Bai, X.C. | | Deposit date: | 2021-05-01 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of the activation of c-MET receptor.
Nat Commun, 12, 2021
|
|
4WKR
 
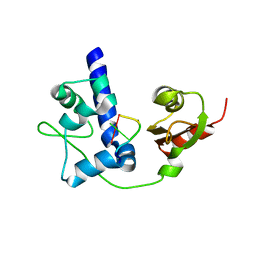 | | LaRP7 wrapping up the 3' hairpin of 7SK non-coding RNA (302-332) | | Descriptor: | 7SK GGHP4 (300-332), La-related protein 7 | | Authors: | Uchikawa, E, Natchiar, K.S, Han, X, Proux, F, Roblin, P, Zhang, E, Durand, A, Klaholz, B.P, Dock-Bregeon, A.-C. | | Deposit date: | 2014-10-03 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insight into the mechanism of stabilization of the 7SK small nuclear RNA by LARP7.
Nucleic Acids Res., 43, 2015
|
|
7MOB
 
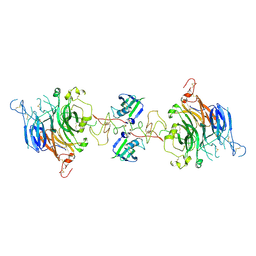 | | Cryo-EM structure of 2:2 c-MET/NK1 complex | | Descriptor: | Hepatocyte growth factor, Hepatocyte growth factor receptor | | Authors: | Uchikawa, E, Chen, Z.M, Xiao, G.Y, Zhang, X.W, Bai, X.C. | | Deposit date: | 2021-05-01 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural basis of the activation of c-MET receptor.
Nat Commun, 12, 2021
|
|
7MO8
 
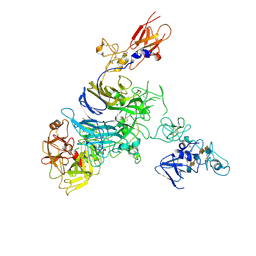 | | Cryo-EM structure of 1:1 c-MET I/HGF I complex after focused 3D refinement of holo-complex | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Hepatocyte growth factor, Hepatocyte growth factor receptor | | Authors: | Uchikawa, E, Chen, Z.M, Xiao, G.Y, Zhang, X.W, Bai, X.C. | | Deposit date: | 2021-05-01 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural basis of the activation of c-MET receptor.
Nat Commun, 12, 2021
|
|
6PXV
 
 | | Cryo-EM structure of full-length insulin receptor bound to 4 insulin. 3D refinement was focused on the extracellular region. | | Descriptor: | Insulin, Insulin receptor | | Authors: | Uchikawa, E, Choi, E, Shang, G.J, Yu, H.T, Bai, X.C. | | Deposit date: | 2019-07-27 | | Release date: | 2019-09-04 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Activation mechanism of the insulin receptor revealed by cryo-EM structure of the fully liganded receptor-ligand complex.
Elife, 8, 2019
|
|
6PXW
 
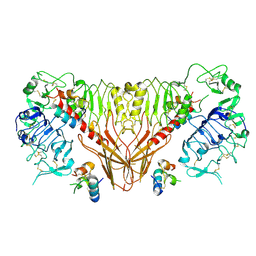 | | Cryo-EM structure of full-length insulin receptor bound to 4 insulin. 3D refinement was focused on the top part of the receptor complex. | | Descriptor: | Insulin, Insulin receptor | | Authors: | Uchikawa, E, Choi, E, Shang, G.J, Yu, H.T, Bai, X.C. | | Deposit date: | 2019-07-28 | | Release date: | 2019-09-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Activation mechanism of the insulin receptor revealed by cryo-EM structure of the fully liganded receptor-ligand complex.
Elife, 8, 2019
|
|
2ZOC
 
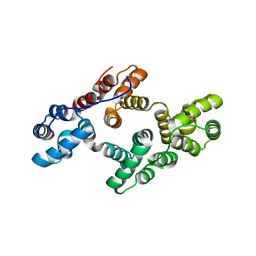 | | Crystal structure of recombinant human annexin IV | | Descriptor: | Annexin A4, CALCIUM ION | | Authors: | Konno, M, Kaneko-Kanzaki, Y, Fushinobu-Okushi, N, Mochizuki, K, Uchikaw, E, Satoh, A, Aikawa, K. | | Deposit date: | 2008-05-08 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The comparison of the loop structure of membrane binding sites between human and bovine annexin IV
To be Published
|
|
5JCH
 
 | |
5JBG
 
 | |
5JC3
 
 | |
5JCF
 
 | |
5JC7
 
 | |
5JBJ
 
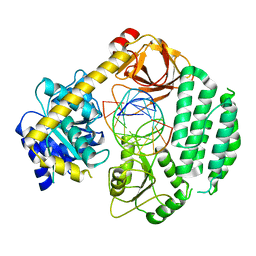 | |
8SBD
 
 | | Cryo-EM structure of insulin amyloid-like fibril that is composed of two antiparallel protofilaments | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Wang, L.W, Hall, C, Uchikawa, E, Chen, D.L, Choi, E, Zhang, X.W, Bai, X.C. | | Deposit date: | 2023-04-03 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of insulin fibrillation.
Sci Adv, 9, 2023
|
|
6VXK
 
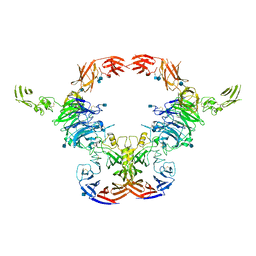 | | Cryo-EM Structure of the full-length A39R/PlexinC1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Plexin-C1, Semaphorin-like protein 139 | | Authors: | Kuo, Y.-C, Chen, H, Shang, G, Uchikawa, E, Tian, H, Bai, X, Zhang, X. | | Deposit date: | 2020-02-22 | | Release date: | 2020-04-29 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of the PlexinC1/A39R complex reveals inter-domain interactions critical for ligand-induced activation.
Nat Commun, 11, 2020
|
|
5JB2
 
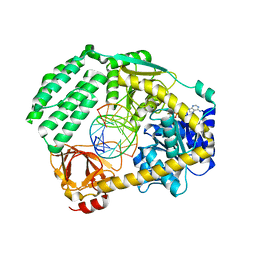 | |
5JAJ
 
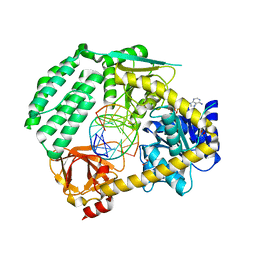 | |
8AQS
 
 | | BA.4/5 SARS-CoV-2 Spike bound to human ACE2 (local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein,Fibritin, ... | | Authors: | Lau, K, Ni, D, Beckert, B, Nazarov, S, Myasnikov, A, Pojer, F, Stahlberg, H, Uchikawa, E. | | Deposit date: | 2022-08-13 | | Release date: | 2023-03-01 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Cryo-EM structures and binding of mouse and human ACE2 to SARS-CoV-2 variants of concern indicate that mutations enabling immune escape could expand host range.
Plos Pathog., 19, 2023
|
|
3VU8
 
 | | Metionyl-tRNA synthetase from Thermus thermophilus complexed with methionyl-adenylate analogue | | Descriptor: | Methionine--tRNA ligase, N-[METHIONYL]-N'-[ADENOSYL]-DIAMINOSULFONE, ZINC ION | | Authors: | Konno, M, Kato-Murayama, M, Toma-Fukai, S, Uchikawa, E, Nureki, O, Yokoyama, S. | | Deposit date: | 2012-06-22 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The modeling of structures of specific conformation of homosysteine-AMP leading to thiolactone-formation on class Ia aminoacyl-tRNA synthetases
To be Published
|
|
8AQT
 
 | | Beta SARS-CoV-2 Spike bound to mouse ACE2 (local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2,Ig gamma-2A chain C region, ... | | Authors: | Lau, K, Ni, D, Beckert, B, Nazarov, S, Myasnikov, A, Pojer, F, Stahlberg, H, Uchikawa, E. | | Deposit date: | 2022-08-13 | | Release date: | 2023-03-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM structures and binding of mouse and human ACE2 to SARS-CoV-2 variants of concern indicate that mutations enabling immune escape could expand host range.
Plos Pathog., 19, 2023
|
|
8AQU
 
 | | BA.1 SARS-CoV-2 Spike bound to mouse ACE2 (local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2,Ig gamma-2A chain C region, ... | | Authors: | Lau, K, Ni, D, Beckert, B, Nazarov, S, Myasnikov, A, Pojer, F, Stahlberg, H, Uchikawa, E. | | Deposit date: | 2022-08-13 | | Release date: | 2023-03-01 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Cryo-EM structures and binding of mouse and human ACE2 to SARS-CoV-2 variants of concern indicate that mutations enabling immune escape could expand host range.
Plos Pathog., 19, 2023
|
|
8AQV
 
 | | BA.2.12.1 SARS-CoV-2 Spike bound to mouse ACE2 (local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2,Ig gamma-2A chain C region, ... | | Authors: | Lau, K, Ni, D, Beckert, B, Nazarov, S, Myasnikov, A, Pojer, F, Stahlberg, H, Uchikawa, E. | | Deposit date: | 2022-08-13 | | Release date: | 2023-03-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Cryo-EM structures and binding of mouse and human ACE2 to SARS-CoV-2 variants of concern indicate that mutations enabling immune escape could expand host range.
Plos Pathog., 19, 2023
|
|
8AQW
 
 | | BA.4/5 SARS-CoV-2 Spike bound to mouse ACE2 (local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2,Ig gamma-2A chain C region, ... | | Authors: | Lau, K, Ni, D, Beckert, B, Nazarov, S, Myasnikov, A, Pojer, F, Stahlberg, H, Uchikawa, E. | | Deposit date: | 2022-08-13 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures and binding of mouse and human ACE2 to SARS-CoV-2 variants of concern indicate that mutations enabling immune escape could expand host range.
Plos Pathog., 19, 2023
|
|
