1V73
 
 | | Crystal Structure of Cold-Active Protein-Tyrosine Phosphatase of a Psychrophile Shewanella SP. | | Descriptor: | ACETIC ACID, CALCIUM ION, psychrophilic phosphatase I | | Authors: | Tsuruta, H, Mikami, B, Aizono, Y. | | Deposit date: | 2003-12-09 | | Release date: | 2005-03-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal Structure of Cold-Active Protein-Tyrosine Phosphatase from a Psychrophile, Shewanella sp
J.Biochem.(Tokyo), 137, 2005
|
|
2Z72
 
 | | New Structure Of Cold-Active Protein Tyrosine Phosphatase At 1.1 Angstrom | | Descriptor: | Protein-tyrosine-phosphatase, ZINC ION | | Authors: | Tsuruta, H, Mikami, B, Yamamoto, C, Yamagata, H. | | Deposit date: | 2007-08-10 | | Release date: | 2008-07-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The role of group bulkiness in the catalytic activity of psychrophile cold-active protein tyrosine phosphatase
Febs J., 275, 2008
|
|
2ZBM
 
 | | Crystal Structure of I115M Mutant Cold-Active Protein Tyrosine Phosphatase | | Descriptor: | Protein-tyrosine-phosphatase, ZINC ION | | Authors: | Tsuruta, H, Mikami, B, Yamamoto, C, Yamagata, H. | | Deposit date: | 2007-10-24 | | Release date: | 2008-07-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The role of group bulkiness in the catalytic activity of psychrophile cold-active protein tyrosine phosphatase
Febs J., 275, 2008
|
|
3A52
 
 | | Crystal structure of cold-active alkailne phosphatase from psychrophile Shewanella sp. | | Descriptor: | Cold-active alkaline phosphatase, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Tsuruta, H, Mikami, B, Higashi, T, Aizono, Y. | | Deposit date: | 2009-07-24 | | Release date: | 2010-04-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of cold-active alkaline phosphatase from the psychrophile Shewanella sp.
Biosci.Biotechnol.Biochem., 74, 2010
|
|
1G3I
 
 | | CRYSTAL STRUCTURE OF THE HSLUV PROTEASE-CHAPERONE COMPLEX | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-DEPENDENT HSLU PROTEASE ATP-BINDING SUBUNIT HSLU, ATP-DEPENDENT PROTEASE HSLV | | Authors: | Sousa, M.C, Trame, C.B, Tsuruta, H, Wilbanks, S.M, Reddy, V.S, McKay, D.B. | | Deposit date: | 2000-10-24 | | Release date: | 2000-11-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Crystal and solution structures of an HslUV protease-chaperone complex.
Cell(Cambridge,Mass.), 103, 2000
|
|
5B1Q
 
 | | Human herpesvirus 6B tegument protein U14 | | Descriptor: | GLYCEROL, U14 protein | | Authors: | Wang, B, Nishimura, M, Tang, H, Kawabata, A, Mahmoud, N.F, Khanlari, Z, Hamada, D, Tsuruta, H, Mori, Y. | | Deposit date: | 2015-12-11 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Human Herpesvirus 6B Tegument Protein U14.
Plos Pathog., 12, 2016
|
|
1SKU
 
 | | E. coli Aspartate Transcarbamylase 240's Loop Mutant (K244N) | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, MALONATE ION, ... | | Authors: | Alam, N, Stieglitz, K.A, Caban, M.D, Gourinath, S, Tsuruta, H, Kantrowitz, E.R. | | Deposit date: | 2004-03-05 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 240s Loop Interactions Stabilize the T State of Escherichia coli Aspartate Transcarbamoylase.
J.Biol.Chem., 279, 2004
|
|
2A0F
 
 | | Structure of D236A mutant E. coli Aspartate Transcarbamoylase in presence of Phosphonoacetamide at 2.90 A resolution | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, PHOSPHONOACETAMIDE, ... | | Authors: | Stieglitz, K.A, Dusinberre, K.J, Cardia, J.P, Tsuruta, H, Kantrowitz, E.R. | | Deposit date: | 2005-06-16 | | Release date: | 2005-09-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the E.coli Aspartate Transcarbamoylase Trapped in the Middle of the Catalytic Cycle.
J.Mol.Biol., 352, 2005
|
|
1XJW
 
 | | The Structure of E. coli Aspartate Transcarbamoylase Q137A Mutant in The R-State | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, N-(PHOSPHONACETYL)-L-ASPARTIC ACID, ... | | Authors: | Stieglitz, K.A, Alam, N, Xia, J, Gourinath, S, Tsuruta, H, Kantrowitz, E.R. | | Deposit date: | 2004-09-25 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | A Single Amino Acid Substitution in the Active Site of Escherichia coli Aspartate Transcarbamoylase Prevents the Allosteric Transition.
J.Mol.Biol., 349, 2005
|
|
2H3E
 
 | | Structure of wild-type E. coli Aspartate Transcarbamoylase in the presence of N-phosphonacetyl-L-isoasparagine at 2.3A resolution | | Descriptor: | (S)-4-AMINO-4-OXO-3-(2-PHOSPHONOACETAMIDO)BUTANOIC ACID, Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, ... | | Authors: | Eldo, J, Cardia, J.P, O'Day, E.M, Xia, J, Tsuruta, H, Kantrowitz, E.R. | | Deposit date: | 2006-05-22 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | N-Phosphonacetyl-l-isoasparagine a Potent and Specific Inhibitor of Escherichia coli Aspartate Transcarbamoylase.
J.Med.Chem., 49, 2006
|
|
2IPO
 
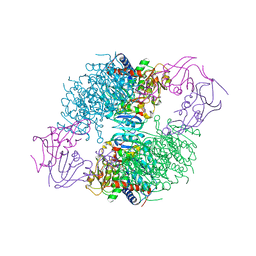 | | E. coli Aspartate Transcarbamoylase complexed with N-phosphonacetyl-L-asparagine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, ... | | Authors: | Cardia, J.P, Eldo, J, Xia, J, O'Day, E.M, Tsuruta, H, Kantrowitz, E.R. | | Deposit date: | 2006-10-12 | | Release date: | 2007-08-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Use of L-asparagine and N-phosphonacetyl-L-asparagine to investigate the linkage of catalysis and homotropic cooperativity in E. coli aspartate transcarbomoylase.
Proteins, 71, 2008
|
|
3DMD
 
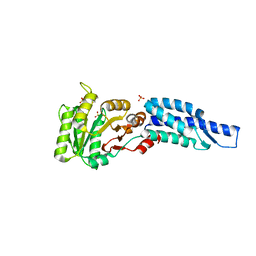 | | Structures and Conformations in Solution of the Signal Recognition Particle Receptor from the archaeon Pyrococcus furiosus | | Descriptor: | GLYCEROL, SULFATE ION, Signal recognition particle receptor | | Authors: | Egea, P.F, Tsuruta, H, Napetschnig, J, Walter, P, Stroud, R.M. | | Deposit date: | 2008-06-30 | | Release date: | 2008-11-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structures of the Signal Recognition Particle Receptor from the Archaeon Pyrococcus furiosus: Implications for the Targeting Step at the Membrane.
Plos One, 3, 2008
|
|
3E70
 
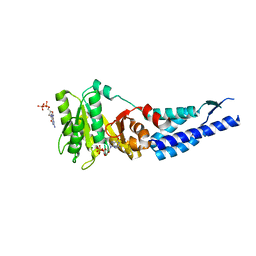 | | Structures and conformations in solution of the Signal Recognition Particle Receptor from the Archaeon Pyrococcus Furiosus | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Signal recognition particle receptor | | Authors: | Egea, P.F, Tsuruta, H, Napetschnig, J, Walter, P, Stroud, R.M. | | Deposit date: | 2008-08-17 | | Release date: | 2008-11-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structures of the signal recognition particle receptor from the archaeon Pyrococcus furiosus: implications for the targeting step at the membrane.
Plos One, 3, 2008
|
|
3DY5
 
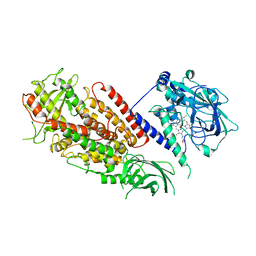 | | Allene oxide synthase 8R-lipoxygenase from Plexaura homomalla | | Descriptor: | Allene oxide synthase-lipoxygenase protein, FE (II) ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gilbert, N.C, Niebuhr, M, Tsuruta, H, Newcomer, M.E. | | Deposit date: | 2008-07-25 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | A covalent linker allows for membrane targeting of an oxylipin biosynthetic complex.
Biochemistry, 47, 2008
|
|
3DM9
 
 | | Structures and Conformations in Solution of the Signal Recognition Particle Receptor from the archaeon Pyrococcus furiosus | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, PHOSPHATE ION, Signal recognition particle receptor | | Authors: | Egea, P.F, Tsuruta, H, Napetschnig, J, Walter, P, Stroud, R.M. | | Deposit date: | 2008-06-30 | | Release date: | 2008-11-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of the Signal Recognition Particle Receptor from the Archaeon Pyrococcus furiosus: Implications for the Targeting Step at the Membrane.
Plos One, 3, 2008
|
|
1I5O
 
 | | CRYSTAL STRUCTURE OF MUTANT R105A OF E. COLI ASPARTATE TRANSCARBAMOYLASE | | Descriptor: | ASPARTATE TRANSCARBAMOYLASE CATALYTIC CHAIN, ASPARTATE TRANSCARBAMOYLASE REGULATORY CHAIN, N-(PHOSPHONACETYL)-L-ASPARTIC ACID, ... | | Authors: | Macol, C.P, Tsuruta, H, Stec, B, Kantrowitz, E.R. | | Deposit date: | 2001-02-28 | | Release date: | 2001-05-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Direct structural evidence for a concerted allosteric transition in Escherichia coli aspartate transcarbamoylase.
Nat.Struct.Biol., 8, 2001
|
|
1IN7
 
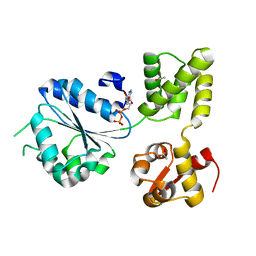 | | THERMOTOGA MARITIMA RUVB R170A | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, HOLLIDAY JUNCTION DNA HELICASE RUVB | | Authors: | Putnam, C.D, Clancy, S.B, Tsuruta, H, Wetmur, J.G, Tainer, J.A. | | Deposit date: | 2001-05-12 | | Release date: | 2001-08-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of the RuvB Holliday junction branch migration motor.
J.Mol.Biol., 311, 2001
|
|
1IN5
 
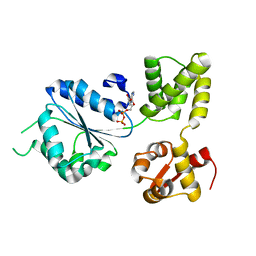 | | THERMOGOTA MARITIMA RUVB A156S MUTANT | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HOLLIDAY JUNCTION DNA HELICASE RUVB | | Authors: | Putnam, C.D, Clancy, S.B, Tsuruta, H, Gonzalez, S, Wetmur, J.G, Tainer, J.A. | | Deposit date: | 2001-05-12 | | Release date: | 2001-08-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and mechanism of the RuvB Holliday junction branch migration motor.
J.Mol.Biol., 311, 2001
|
|
1J7K
 
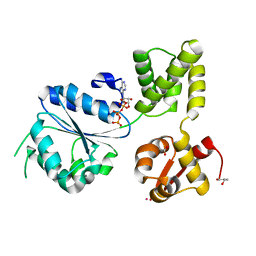 | | THERMOTOGA MARITIMA RUVB P216G MUTANT | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, COBALT (II) ION, ... | | Authors: | Putnam, C.D, Clancy, S.B, Tsuruta, H, Wetmur, J.G, Tainer, J.A. | | Deposit date: | 2001-05-16 | | Release date: | 2001-08-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and mechanism of the RuvB Holliday junction branch migration motor.
J.Mol.Biol., 311, 2001
|
|
1IN8
 
 | | THERMOTOGA MARITIMA RUVB T158V | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HOLLIDAY JUNCTION DNA HELICASE RUVB | | Authors: | Putnam, C.D, Clancy, S.B, Tsuruta, H, Wetmur, J.G, Tainer, J.A. | | Deposit date: | 2001-05-12 | | Release date: | 2001-08-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of the RuvB Holliday junction branch migration motor.
J.Mol.Biol., 311, 2001
|
|
1IN6
 
 | | THERMOTOGA MARITIMA RUVB K64R MUTANT | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, COBALT (II) ION, ... | | Authors: | Putnam, C.D, Clancy, S.B, Tsuruta, H, Wetmur, J.G, Tainer, J.A. | | Deposit date: | 2001-05-12 | | Release date: | 2001-08-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and mechanism of the RuvB Holliday junction branch migration motor.
J.Mol.Biol., 311, 2001
|
|
