2PTQ
 
 | |
2PTR
 
 | |
2PTS
 
 | |
4XQ8
 
 | |
8GQO
 
 | |
1U16
 
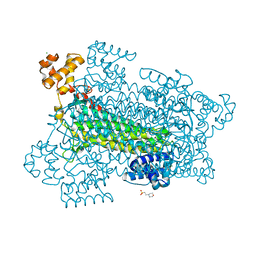 | | Crystal structure of a duck-delta-crystallin-1 double loop mutant (DLM) in complex with sulfate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Delta crystallin I, ... | | Authors: | Tsai, M, Sampaleanu, L.M, Greene, C, Creagh, L, Haynes, C, Howell, P.L. | | Deposit date: | 2004-07-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A duck delta1 crystallin double loop mutant provides insight into residues important for argininosuccinate lyase activity.
Biochemistry, 43, 2004
|
|
1U15
 
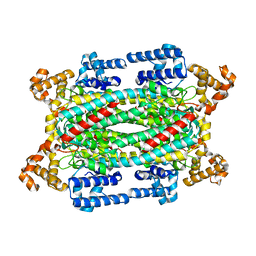 | | Crystal structure of a duck-delta-crystallin-1 double loop mutant (DLM) | | Descriptor: | Delta crystallin I | | Authors: | Tsai, M, Sampaleanu, L.M, Greene, C, Creagh, L, Haynes, C, Howell, P.L. | | Deposit date: | 2004-07-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A duck delta1 crystallin double loop mutant provides insight into residues important for argininosuccinate lyase activity.
Biochemistry, 43, 2004
|
|
7D0V
 
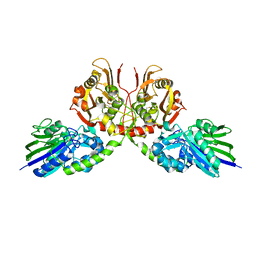 | | Crystal structure of Taiwan cobra 5'-nucleotidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Snake venom 5'-nucleotidase, ... | | Authors: | Tsai, M.-H, Lin, I.-J, Lin, C.-C, Wu, W.-G. | | Deposit date: | 2020-09-12 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.175 Å) | | Cite: | Crystal structure of Taiwan cobra 5'-nucleotidase
To Be Published
|
|
3O7W
 
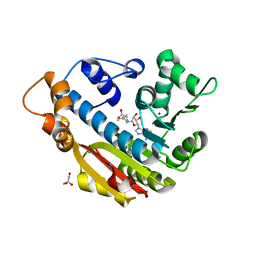 | | The Crystal Structure of Human Leucine Carboxyl Methyltransferase 1 | | Descriptor: | GLYCEROL, Leucine carboxyl methyltransferase 1, S-ADENOSYLMETHIONINE, ... | | Authors: | Tsai, M.L, Cronin, N, Djordjevic, S. | | Deposit date: | 2010-08-01 | | Release date: | 2010-09-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of human leucine carboxyl methyltransferase 1 that regulates protein phosphatase PP2A
Acta Crystallogr.,Sect.D, 67, 2011
|
|
1GH4
 
 | | Structure of the triple mutant (K56M, K120M, K121M) of phospholipase A2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Sekar, K, Velmurugan, D, Tsai, M.D. | | Deposit date: | 2000-11-09 | | Release date: | 2001-05-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Observation of additional calcium ion in the crystal structure of the triple mutant K56,120,121M of bovine pancreatic phospholipase A2.
J.Mol.Biol., 324, 2002
|
|
4ZGI
 
 | | Structure of Truncated Human TIFA | | Descriptor: | TRAF-interacting protein with FHA domain-containing protein A | | Authors: | Weng, J.H, Wei, T.Y.W, Hsieh, Y.C, Huang, C.C.F, Wu, P.Y.G, Chen, E.S.W, Huang, K.F, Chen, C.J, Tsai, M.D. | | Deposit date: | 2015-04-23 | | Release date: | 2015-10-14 | | Last modified: | 2015-10-28 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Uncovering the Mechanism of Forkhead-Associated Domain-Mediated TIFA Oligomerization That Plays a Central Role in Immune Responses.
Biochemistry, 54, 2015
|
|
6LY6
 
 | | PylRS C-terminus domain mutant bound with 3-(1-Naphthyl)-L-alanine and AMPNP | | Descriptor: | MAGNESIUM ION, NAPHTHALEN-2-YL-3-ALANINE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Weng, J.H, Tsai, M.D, Wang, Y.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5001328 Å) | | Cite: | Probing the Active Site of Deubiquitinase USP30 with Noncanonical Tryptophan Analogues.
Biochemistry, 59, 2020
|
|
6LY3
 
 | | PylRS C-terminus domain mutant bound with 3-Benzothienyl-L-alanine and AMPNP | | Descriptor: | 3-(1-benzothiophen-3-yl)-L-alanine, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Weng, J.H, Tsai, M.D, Wang, Y.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89590144 Å) | | Cite: | Probing the Active Site of Deubiquitinase USP30 with Noncanonical Tryptophan Analogues.
Biochemistry, 59, 2020
|
|
1A5E
 
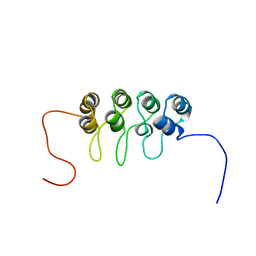 | | SOLUTION NMR STRUCTURE OF TUMOR SUPPRESSOR P16INK4A, 18 STRUCTURES | | Descriptor: | TUMOR SUPPRESSOR P16INK4A | | Authors: | Byeon, I.-J.L, Li, J, Ericson, K, Selby, T.L, Tevelev, A, Kim, H.-J, O'Maille, P, Tsai, M.-D. | | Deposit date: | 1998-02-13 | | Release date: | 1999-08-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Tumor suppressor p16INK4A: determination of solution structure and analyses of its interaction with cyclin-dependent kinase 4.
Mol.Cell, 1, 1998
|
|
8XMI
 
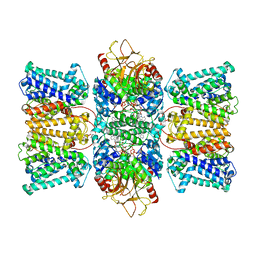 | | Potassium transporter KtrAB from Bacillus subtilis in ATP-bound state with addition of EDTA and EGTA, C1 symmetry | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Ktr system potassium uptake protein A, Ktr system potassium uptake protein B, ... | | Authors: | Chang, Y.K, Chiang, W.T, Hu, N.J, Tsai, M.D. | | Deposit date: | 2023-12-27 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis and synergism of ATP and Na + activation in bacterial K + uptake system KtrAB.
Nat Commun, 15, 2024
|
|
8XMH
 
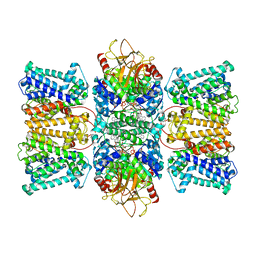 | | Potassium transporter KtrAB from Bacillus subtilis in ATP-bound state with addition of EDTA and EGTA, vertical C2 symmetry axis | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Ktr system potassium uptake protein A, Ktr system potassium uptake protein B, ... | | Authors: | Chang, Y.K, Chiang, W.T, Hu, N.J, Tsai, M.D. | | Deposit date: | 2023-12-27 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis and synergism of ATP and Na + activation in bacterial K + uptake system KtrAB.
Nat Commun, 15, 2024
|
|
5DKW
 
 | |
8IO9
 
 | |
8IO6
 
 | |
8IOE
 
 | |
5ZM0
 
 | | X-ray structure of animal-like Cryptochrome from Chlamydomonas reinhardtii | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Cryptochrome photoreceptor, ... | | Authors: | Franz, S, Ignatz, E, Wenzel, S, Zielosko, H, Gusti Ngurah Putu, E.P, Maestre-Reyna, M, Tsai, M.-D, Yamamoto, J, Mittag, M, Essen, L.-O. | | Deposit date: | 2018-03-31 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the bifunctional cryptochrome aCRY from Chlamydomonas reinhardtii
Nucleic Acids Res., 46, 2018
|
|
8IOA
 
 | |
8IO7
 
 | |
8IO8
 
 | |
5CWR
 
 | |
