3MY2
 
 | |
2NTM
 
 | |
2NTK
 
 | | Crystal structure of PurO/IMP from Methanothermobacter thermoautotrophicus | | Descriptor: | IMP cyclohydrolase, INOSINIC ACID | | Authors: | Kang, Y.N, Tran, A, White, R.H, Ealick, S.E. | | Deposit date: | 2006-11-07 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A novel function for the N-terminal nucleophile hydrolase fold demonstrated by the structure of an archaeal inosine monophosphate cyclohydrolase.
Biochemistry, 46, 2007
|
|
2NTL
 
 | | Crystal structure of PurO/AICAR from Methanothermobacter thermoautotrophicus | | Descriptor: | AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, IMP cyclohydrolase | | Authors: | Kang, Y.N, Tran, A, White, R.H, Ealick, S.E. | | Deposit date: | 2006-11-07 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A novel function for the N-terminal nucleophile hydrolase fold demonstrated by the structure of an archaeal inosine monophosphate cyclohydrolase.
Biochemistry, 46, 2007
|
|
3MV1
 
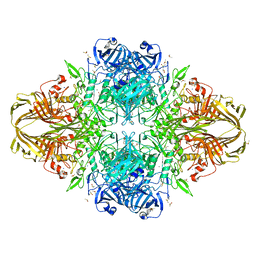 | | E.Coli (lacZ) beta-galactosidase (R599A) in complex with Guanidinium | | Descriptor: | Beta-galactosidase, DIMETHYL SULFOXIDE, GUANIDINE, ... | | Authors: | Dugdale, M.L, Vance, M, Driedger, M.L, Nibber, A, Tran, A, Huber, R.E. | | Deposit date: | 2010-05-03 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Importance of Arg-599 of b-galactosidase (Escherichia coli) as an anchor for the open conformations of Phe-601 and the active-site loop
Biochem.Cell Biol., 88, 2010
|
|
3MUZ
 
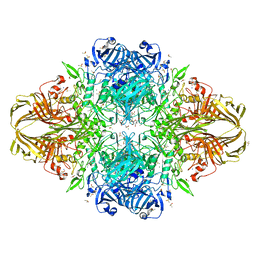 | | E.Coli (lacZ) beta-galactosidase (R599A) in complex with IPTG | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Dugdale, M.L, Vance, M.L, Driedger, M.R, Nibber, A, Tran, A, Huber, R.E. | | Deposit date: | 2010-05-03 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Importance of Arg-599 of b-galactosidase (Escherichia coli) as an anchor for the open conformations of Phe-601 and the active-site loop
Biochem.Cell Biol., 88, 2010
|
|
3MUY
 
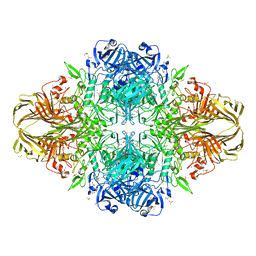 | | E. coli (lacZ) beta-galactosidase (R599A) | | Descriptor: | Beta-D-galactosidase, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Dugdale, M.L, Vance, M, Driedger, M.L, Nibber, A, Tran, A, Huber, R.E. | | Deposit date: | 2010-05-03 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Importance of Arg-599 of beta-galactosidase (Escherichia coli) as an anchor for the open conformations of Phe-601 and the active-site loop
Biochem.Cell Biol., 88, 2010
|
|
3MV0
 
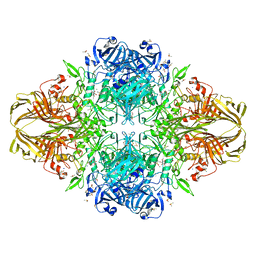 | | E. COLI (lacZ) BETA-GALACTOSIDASE (R599A) IN COMPLEX WITH D-GALCTOPYRANOSYL-1-ONE | | Descriptor: | Beta-galactosidase, D-galactonolactone, DIMETHYL SULFOXIDE, ... | | Authors: | Dugdale, M.L, Vance, M, Driedger, M.L, Nibber, A, Tran, A, Huber, R.E. | | Deposit date: | 2010-05-03 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Importance of Arg-599 of b-galactosidase (Escherichia coli) as an anchor for the open conformations of Phe-601 and the active-site loop
Biochem.Cell Biol., 88, 2010
|
|
3P50
 
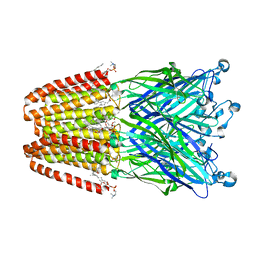 | | Structure of propofol bound to a pentameric ligand-gated ion channel, GLIC | | Descriptor: | 2,6-BIS(1-METHYLETHYL)PHENOL, DIUNDECYL PHOSPHATIDYL CHOLINE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Nury, H, Van Renterghem, C, Weng, Y, Tran, A, Baaden, M, Dufresne, V, Changeux, J.P, Sonner, J.M, Delarue, M, Corringer, P.J. | | Deposit date: | 2010-10-07 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | X-ray structures of general anaesthetics bound to a pentameric ligand-gated ion channel
Nature, 469, 2011
|
|
3P4W
 
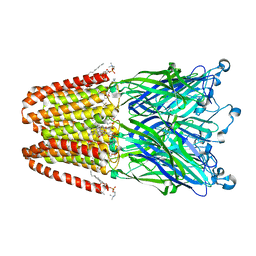 | | Structure of desflurane bound to a pentameric ligand-gated ion channel, GLIC | | Descriptor: | (2S)-2-(difluoromethoxy)-1,1,1,2-tetrafluoroethane, DIUNDECYL PHOSPHATIDYL CHOLINE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Nury, H, Van Renterghem, C, Weng, Y, Tran, A, Baaden, M, Dufresne, V, Changeux, J.P, Sonner, J.M, Delarue, M, Corringer, P.J. | | Deposit date: | 2010-10-07 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray structures of general anaesthetics bound to a pentameric ligand-gated ion channel
Nature, 469, 2011
|
|
4Q9T
 
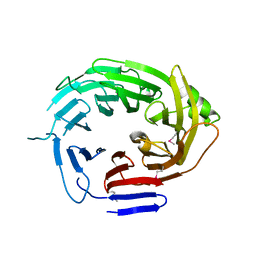 | |
4JLQ
 
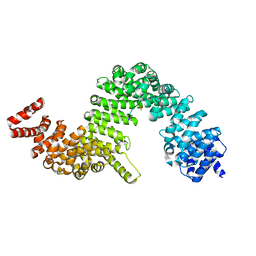 | | Crystal structure of human Karyopherin-beta2 bound to the PY-NLS of Saccharomyces cerevisiae NAB2 | | Descriptor: | Nuclear polyadenylated RNA-binding protein NAB2, Transportin-1 | | Authors: | Sampathkumar, P, Gizzi, A, Rout, M.P, Chook, Y.M, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Nucleocytoplasmic Transport: a Target for Cellular Control (NPCXstals) | | Deposit date: | 2013-03-12 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of human Karyopherin beta 2 bound to the PY-NLS of Saccharomyces cerevisiae Nab2.
J.Struct.Funct.Genom., 14, 2013
|
|
4OO6
 
 | | Crystal structure of human KAP-beta2 bound to the NLS of HCC1 (Hepato Cellular Carcinoma protein 1) | | Descriptor: | RNA-binding protein 39, Transportin-1 | | Authors: | Sampathkumar, P, Brower, A, Soniat, M, Bonanno, J, Hillerich, B, Seidel, R.D, Rout, M.P, Chook, Y.M, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Nucleocytoplasmic Transport: a Target for Cellular Control (NPCXstals) | | Deposit date: | 2014-01-30 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of human KAP-beta2 bound to the NLS of HCC1 (Hepato Cellular Carcinoma protein 1)
to be published
|
|
6LKT
 
 | | Crystal structure of the Fab fragment of murine monoclonal antibody KH-1 against Human herpesvirus 6B | | Descriptor: | antibody Fab Fragment L-chain, antibody Fab fragment H chain | | Authors: | Nishimura, M, Novita, B.D, Kato, T, Tjan, L.H, Wang, B, Wakata, A, Poetranto, A.L, Kawabata, A, Tang, H, Aoshi, T, Mori, Y. | | Deposit date: | 2019-12-20 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the interaction of human herpesvirus 6B tetrameric glycoprotein complex with the cellular receptor, human CD134.
Plos Pathog., 16, 2020
|
|
6LTG
 
 | | Crystal structure of the Fab fragment of murine monoclonal antibody OHV-3 against Human herpesvirus 6B | | Descriptor: | MAGNESIUM ION, antibody Fab fragment H-chain, antibody Fab fragment L-chain | | Authors: | Nishimura, M, Novita, B.D, Kato, T, Tjan, L.H, Wang, B, Wakata, A, Poetranto, A.L, Kawabata, A, Tang, H, Aoshi, T, Mori, Y. | | Deposit date: | 2020-01-22 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural basis for the interaction of human herpesvirus 6B tetrameric glycoprotein complex with the cellular receptor, human CD134.
Plos Pathog., 16, 2020
|
|
4IFQ
 
 | | Crystal structure of Saccharomyces cerevisiae NUP192, residues 2 to 960 [ScNup192(2-960)] | | Descriptor: | IODIDE ION, Nucleoporin NUP192, SULFATE ION | | Authors: | Sampathkumar, P, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Nucleocytoplasmic Transport: a Target for Cellular Control (NPCXstals) | | Deposit date: | 2012-12-14 | | Release date: | 2013-02-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure, dynamics, evolution, and function of a major scaffold component in the nuclear pore complex.
Structure, 21, 2013
|
|
2M4M
 
 | | Solution structure of the RRM domain of the hypothetical protein CAGL0M09691g from Candida glabrata | | Descriptor: | hypothetical protein | | Authors: | Harris, R, Hillerich, B, Ahmed, M, Bonanno, J.B, Chamala, S, Evans, B, Lafleur, J, Hammonds, J, Washington, E, Stead, M, Love, J, Attonito, J, Patel, H, Seidel, R.D, Chook, Y.M, Rout, M.P, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Nucleocytoplasmic Transport: a Target for Cellular Control (NPCXstals) | | Deposit date: | 2013-02-07 | | Release date: | 2013-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RRM domain of the hypothetical protein CAGL0M09691g from Candida glabrata
To be Published
|
|
3REV
 
 | | Crystal structure of human alloreactive tcr nb20 | | Descriptor: | 1,2-ETHANEDIOL, TCR NB20 ALPHA CHAIN, TCR NB20 BETA CHAIN | | Authors: | Wood, A, Mohammed, F, Salim, M, Tranter, A, Rickinson, A.B, Moss, P.A.H, Stauss, H.J, Steven, N.M, Willcox, B.E. | | Deposit date: | 2011-04-05 | | Release date: | 2012-01-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and energetic evidence for highly peptide-specific tumor antigen targeting via allo-MHC restriction.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3REW
 
 | | Crystal structure of an lmp2a-derived peptide bound to human class i mhc hla-a2 | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, CHLORIDE ION, ... | | Authors: | Wood, A, Mohammed, F, Salim, M, Tranter, A, Rickinson, A.B, Moss, P.A.H, Stauss, H.J, Steven, N.M, Willcox, B.E. | | Deposit date: | 2011-04-05 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and energetic evidence for highly peptide-specific tumor antigen targeting via allo-MHC restriction.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
