3QLK
 
 | | Crystal Structure of RipA from Yersinia pestis | | Descriptor: | Coenzyme A transferase | | Authors: | Torres, R, Goulding, C.W. | | Deposit date: | 2011-02-02 | | Release date: | 2012-01-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Biochemical, structural and molecular dynamics analyses of the potential virulence factor RipA from Yersinia pestis.
Plos One, 6, 2011
|
|
3S8D
 
 | |
3QLI
 
 | | Crystal Structure of RipA from Yersinia pestis | | Descriptor: | ACETATE ION, Coenzyme A transferase | | Authors: | Torres, R, Goulding, C.W. | | Deposit date: | 2011-02-02 | | Release date: | 2012-01-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical, structural and molecular dynamics analyses of the potential virulence factor RipA from Yersinia pestis.
Plos One, 6, 2011
|
|
4Y0L
 
 | | Mycobacterial membrane protein MmpL11D2 | | Descriptor: | IODIDE ION, Putative membrane protein mmpL11, SULFATE ION | | Authors: | Torres, R, Chim, N, Goulding, C.W. | | Deposit date: | 2015-02-06 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Structure and Interactions of Periplasmic Domains of Crucial MmpL Membrane Proteins from Mycobacterium tuberculosis.
Chem.Biol., 22, 2015
|
|
4YMP
 
 | |
3QLL
 
 | | Crystal Structure of RipC from Yersinia pestis | | Descriptor: | Citrate lyase | | Authors: | Torres, R, Goulding, C.W. | | Deposit date: | 2011-02-02 | | Release date: | 2012-01-11 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural insights into RipC, a putative citrate lyase beta subunit from a Yersinia pestis virulence operon
Acta Crystallogr.,Sect.F, 68, 2012
|
|
4LPN
 
 | | Frog M-ferritin with cobalt, D127E mutant | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Ferritin, ... | | Authors: | Torres, R, Behera, R, Goulding, C.W. | | Deposit date: | 2013-07-16 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | D127E ion channel exit modification in ferritin nanocages entraps Fe(II) and impairs its distribution to diiron catalytic centers
To be Published
|
|
4LPM
 
 | | Frog M-ferritin with magnesium, D127E mutant | | Descriptor: | CHLORIDE ION, Ferritin, middle subunit, ... | | Authors: | Torres, R, Behera, R, Goulding, C.W, Theil, E.C. | | Deposit date: | 2013-07-16 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | D127E ion channel exit modification in ferritin nanocages entraps Fe(II) and impairs its distribution to diiron catalytic centers
To be Published
|
|
4N8J
 
 | | F60M mutant, RipA structure | | Descriptor: | ACETATE ION, COENZYME A, PHOSPHATE ION, ... | | Authors: | Torres, R, Goulding, C.W. | | Deposit date: | 2013-10-17 | | Release date: | 2014-04-09 | | Last modified: | 2014-04-16 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural snapshots along the reaction pathway of Yersinia pestis RipA, a putative butyryl-CoA transferase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4N8K
 
 | | E249A mutant, RipA structure | | Descriptor: | Putative 4-hydroxybutyrate coenzyme A transferase | | Authors: | Torres, R, Goulding, C.W. | | Deposit date: | 2013-10-17 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural snapshots along the reaction pathway of Yersinia pestis RipA, a putative butyryl-CoA transferase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4N8I
 
 | | M31G mutant, RipA structure | | Descriptor: | 4-hydroxybutyrate coenzyme A transferase, ACETATE ION, COENZYME A | | Authors: | Torres, R, Goulding, C.W. | | Deposit date: | 2013-10-17 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Structural snapshots along the reaction pathway of Yersinia pestis RipA, a putative butyryl-CoA transferase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4N8L
 
 | | E249D mutant, RipA structure | | Descriptor: | Putative 4-hydroxybutyrate coenzyme A transferase | | Authors: | Torres, R, Goulding, C.W. | | Deposit date: | 2013-10-17 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.817 Å) | | Cite: | Structural snapshots along the reaction pathway of Yersinia pestis RipA, a putative butyryl-CoA transferase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4N8H
 
 | | E61V mutant, RipA structure | | Descriptor: | 4-hydroxybutyrate coenzyme A transferase | | Authors: | Torres, R, Goulding, C.W. | | Deposit date: | 2013-10-17 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural snapshots along the reaction pathway of Yersinia pestis RipA, a putative butyryl-CoA transferase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5FM2
 
 | | Crystal structure of hyper-phosphorylated RET kinase domain with (proximal) juxtamembrane segment | | Descriptor: | 1-TER-BUTYL-3-P-TOLYL-1H-PYRAZOLO[3,4-D]PYRIMIDIN-4-YLAMINE, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE RECEPTOR RET | | Authors: | Plaza-Menacho, I, Barnouin, K, Barry, R, Borg, A, Orme, M, Mouilleron, S, Martinez-Torres, R.J, Meier, P, McDonald, N.Q. | | Deposit date: | 2015-10-30 | | Release date: | 2016-12-28 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | RET Functions as a Dual-Specificity Kinase that Requires Allosteric Inputs from Juxtamembrane Elements.
Cell Rep, 17, 2016
|
|
5FM3
 
 | | Crystal structure of hyper-phosphorylated RET kinase domain with (proximal) juxtamembrane segment | | Descriptor: | 1-TER-BUTYL-3-P-TOLYL-1H-PYRAZOLO[3,4-D]PYRIMIDIN-4-YLAMINE, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE RECEPTOR RET | | Authors: | Plaza-Menacho, I, Barnouin, K, Barry, R, Borg, A, Orme, M, Mouilleron, S, Martinez-Torres, R.J, Meier, P, McDonald, N.Q. | | Deposit date: | 2015-10-30 | | Release date: | 2016-12-28 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | RET Functions as a Dual-Specificity Kinase that Requires Allosteric Inputs from Juxtamembrane Elements.
Cell Rep, 17, 2016
|
|
5C23
 
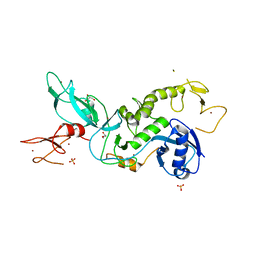 | | Parkin (S65DUblR0RBR) | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase parkin, GLYCEROL, ... | | Authors: | Kumar, A, Aguirre, J.D, Condos, T.E.C, Martinez-Torres, R.J, Chaugule, V.K, Toth, R, Sundaramoorthy, R, Mercier, P, Knebel, A, Spratt, D.E, Barber, K.R, Shaw, G.S, Walden, H. | | Deposit date: | 2015-06-15 | | Release date: | 2015-07-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Disruption of the autoinhibited state primes the E3 ligase parkin for activation and catalysis.
Embo J., 34, 2015
|
|
5C1Z
 
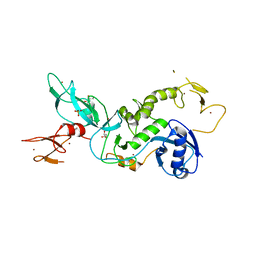 | | Parkin (UblR0RBR) | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase parkin, GLYCEROL, ... | | Authors: | kumar, A, Aguirre, J.D, Condos, T.E.C, Martinez-Torres, R.J, Chaugule, V.K, Toth, R, Sundaramoorthy, R, Mercier, P, Knebel, A, Spratt, D.E, Barber, K.R, Shaw, G.S, Walden, H. | | Deposit date: | 2015-06-15 | | Release date: | 2015-07-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Disruption of the autoinhibited state primes the E3 ligase parkin for activation and catalysis.
Embo J., 34, 2015
|
|
4CMF
 
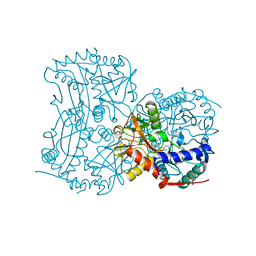 | | The (R)-selective transaminase from Nectria haematococca with inhibitor bound | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-[O-PHOSPHONOPYRIDOXYL]--AMINO-BENZOIC ACID, AMINOTRANSFERASE, ... | | Authors: | Sayer, C, Isupov, M, Martinez-Torres, R.J, Richter, N, Hailes, H.C, Ward, J, Littlechild, J. | | Deposit date: | 2014-01-16 | | Release date: | 2014-03-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Substrate Specificity, Enantioselectivity and Structure of the (R)-Selective Amine:Pyruvate Transaminase from Nectria Haematococca.
FEBS J., 281, 2014
|
|
4CMD
 
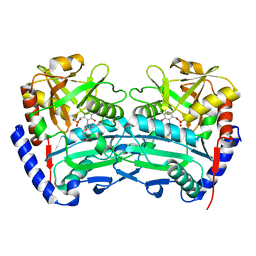 | | The (R)-selective transaminase from Nectria haematococca | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Sayer, C, Isupov, M, Martinez-Torres, R.J, Richter, N, Hailes, H.C, Ward, J, Littlechild, J. | | Deposit date: | 2014-01-16 | | Release date: | 2014-03-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The Substrate Specificity, Enantioselectivity and Structure of the (R)-Selective Amine:Pyruvate Transaminase from Nectria Haematococca.
FEBS J., 281, 2014
|
|
4CKJ
 
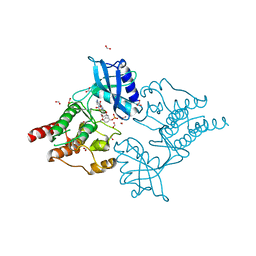 | | Crystal structure of RET tyrosine kinase domain bound to adenosine | | Descriptor: | ADENOSINE, FORMIC ACID, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE RECEPTOR RET | | Authors: | Plaza-Menacho, I, Barnouin, K, Goodman, K, Martinez-Torres, R.J, Borg, A, Murray-Rust, J, Mouilleron, S, Knowles, P, McDonald, N.Q. | | Deposit date: | 2014-01-06 | | Release date: | 2014-03-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Oncogenic RET kinase domain mutations perturb the autophosphorylation trajectory by enhancing substrate presentation in trans.
Mol. Cell, 53, 2014
|
|
4CKI
 
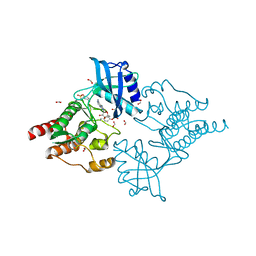 | | Crystal Structure of oncogenic RET tyrosine kinase M918T bound to adenosine | | Descriptor: | ADENOSINE, FORMIC ACID, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE RECEPTOR RET | | Authors: | Plaza-Menacho, I, Barnouin, K, Goodman, K, Martinez-Torres, R.J, Borg, A, Murray-Rust, J, Mouilleron, S, Knowles, P, McDonald, N.Q. | | Deposit date: | 2014-01-06 | | Release date: | 2014-03-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.116 Å) | | Cite: | Oncogenic RET kinase domain mutations perturb the autophosphorylation trajectory by enhancing substrate presentation in trans.
Mol. Cell, 53, 2014
|
|
