1ESV
 
 | | COMPLEX BETWEEN LATRUNCULIN A:RABBIT MUSCLE ALPHA ACTIN:HUMAN GELSOLIN DOMAIN 1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ALPHA ACTIN, CALCIUM ION, ... | | Authors: | Morton, W.M, Ayscough, K.A, McLaughlin, P.J. | | Deposit date: | 2000-04-11 | | Release date: | 2000-07-19 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Latrunculin alters the actin-monomer subunit interface to prevent polymerization.
Nat.Cell Biol., 2, 2000
|
|
1GL2
 
 | | Crystal structure of an endosomal SNARE core complex | | Descriptor: | ENDOBREVIN, SYNTAXIN 7, SYNTAXIN 8, ... | | Authors: | Antonin, W, Becker, S, Jahn, R, Schneider, T.R. | | Deposit date: | 2001-08-22 | | Release date: | 2002-01-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Endosomal Snare Complex Reveals Common Structural Principles of All Snares.
Nat.Struct.Biol., 9, 2001
|
|
1Z3S
 
 | |
1Z3U
 
 | |
1SHW
 
 | | EphB2 / EphrinA5 Complex Structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ephrin type-B receptor 2, Ephrin-A5, ... | | Authors: | Himanen, J.P, Chumley, M.J, Lackmann, M, Li, C, Barton, W.A, Jeffrey, P.D, Vearing, C, Geleick, D, Feldheim, D.A, Boyd, A.W. | | Deposit date: | 2004-02-26 | | Release date: | 2004-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Repelling class discrimination: ephrin-A5 binds to and activates EphB2 receptor signaling
Nat.Neurosci., 7, 2004
|
|
1SHX
 
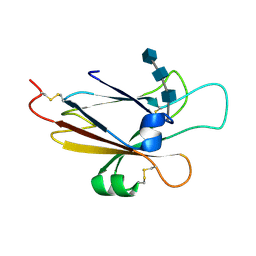 | | Ephrin A5 ligand structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ephrin-A5 | | Authors: | Himanen, J.P, Barton, W.A, Nikolov, D.B, Jeffrey, P.D. | | Deposit date: | 2004-02-26 | | Release date: | 2005-04-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three distinct molecular surfaces in ephrin-A5 are essential for a functional interaction with EphA3.
J.Biol.Chem., 280, 2005
|
|
1SD0
 
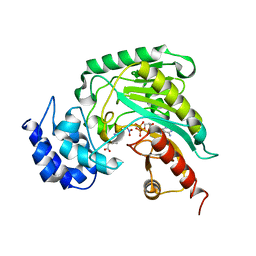 | | Structure of arginine kinase C271A mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ARGININE, Arginine kinase, ... | | Authors: | Gattis, J.L, Ruben, E, Fenley, M.O, Ellington, W.R, Chapman, M.S. | | Deposit date: | 2004-02-12 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The active site cysteine of arginine kinase: structural and functional analysis of partially active mutants
Biochemistry, 43, 2004
|
|
4KGO
 
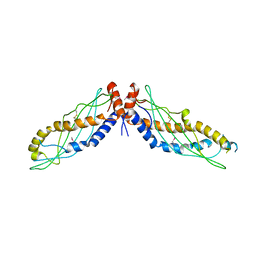 | |
4KGH
 
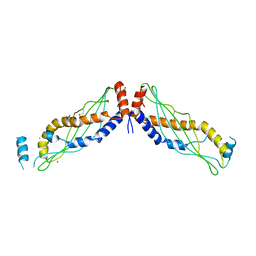 | |
4K59
 
 | |
3M10
 
 | | Substrate-free form of Arginine Kinase | | Descriptor: | Arginine kinase, SULFATE ION | | Authors: | Yousef, M.S, Clark, S.A, Pruett, P.K, Somasundaram, T, Ellington, W.R, Chapman, M.S. | | Deposit date: | 2010-03-03 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.727 Å) | | Cite: | Arginine kinase: joint crystallographic and NMR RDC analyses link substrate-associated motions to intrinsic flexibility.
J.Mol.Biol., 405, 2011
|
|
6BJQ
 
 | |
6BO6
 
 | | Eubacterium eligens beta-glucuronidase bound to UNC4917 glucuronic acid conjugate | | Descriptor: | 4-(4-beta-D-glucopyranuronosylpiperazin-1-yl)-2,7-bis(methylamino)pyrido[3',2':4,5]thieno[3,2-d]pyrimidine, Glycoside Hydrolase Family 2 candidate b-glucuronidase | | Authors: | Pellock, S.J, Walton, W.G, Redinbo, M.R. | | Deposit date: | 2017-11-18 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Gut Microbial beta-Glucuronidase Inhibition via Catalytic Cycle Interception.
ACS Cent Sci, 4, 2018
|
|
6BJW
 
 | |
5A9Q
 
 | | Human nuclear pore complex | | Descriptor: | NUCLEAR PORE COMPLEX PROTEIN NUP107, NUCLEAR PORE COMPLEX PROTEIN NUP133, NUCLEAR PORE COMPLEX PROTEIN NUP155, ... | | Authors: | von Appen, A, Kosinski, J, Sparks, L, Ori, A, DiGuilio, A, Vollmer, B, Mackmull, M, Banterle, N, Parca, L, Kastritis, P, Buczak, K, Mosalaganti, S, Hagen, W, Andres-Pons, A, Lemke, E.A, Bork, P, Antonin, W, Glavy, J.S, Bui, K.H, Beck, M. | | Deposit date: | 2015-07-22 | | Release date: | 2015-09-30 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (23 Å) | | Cite: | In Situ Structural Analysis of the Human Nuclear Pore Complex
Nature, 526, 2015
|
|
5FKI
 
 | | Pseudorabies virus (PrV) nuclear egress complex proteins fitted as a hexameric lattice into a sub-tomogram average derived from focused- ion beam milled lamellae electron cryo-microscopic data | | Descriptor: | CHLORIDE ION, UL31, UL34 protein, ... | | Authors: | Hagen, C, Dent, K.C, Zeev Ben Mordehai, T, Vasishtan, D, Antonin, W, Mettenleiter, T.C, Gruenewald, K. | | Deposit date: | 2015-10-16 | | Release date: | 2016-03-16 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (35 Å) | | Cite: | Crystal Structure of the Herpesvirus Nuclear Egress Complex Provides Insights Into Inner Nuclear Membrane Remodelling
Cell Rep., 13, 2015
|
|
5FDN
 
 | |
6ECA
 
 | | Lactobacillus rhamnosus Beta-glucuronidase | | Descriptor: | Beta-glucuronidase, CHLORIDE ION, GLYCEROL | | Authors: | Biernat, K.A, Pellock, S.J, Bhatt, A.P, Bivins, M.M, Walton, W.G, Tran, B.N.T, Wei, L, Snider, M.C, Cesmat, A.P, Tripathy, A, Erie, D.A, Redinbo, M.R.R. | | Deposit date: | 2018-08-07 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.853 Å) | | Cite: | Structure, function, and inhibition of drug reactivating human gut microbial beta-glucuronidases.
Sci Rep, 9, 2019
|
|
6D4O
 
 | | Eubacterium eligens beta-glucuronidase bound to an amoxapine-glucuronide conjugate | | Descriptor: | (5aR,9aR)-2-chloro-11-(4-beta-D-glucopyranuronosylpiperazin-1-yl)-5a,6,9,9a-tetrahydrodibenzo[b,f][1,4]oxazepine, Beta-glucuronidase, CHLORIDE ION, ... | | Authors: | Pellock, S.J, Walton, W.G, Redinbo, M.R. | | Deposit date: | 2018-04-18 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Gut Microbial beta-Glucuronidase Inhibition via Catalytic Cycle Interception.
ACS Cent Sci, 4, 2018
|
|
7L19
 
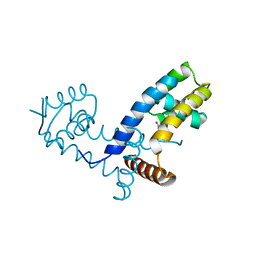 | | Crystal structure of the MarR family transcriptional regulator from Enterobacter soli strain LF7 bound to Indole 3 acetic acid | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, MarR family transcriptional regulator, NICKEL (II) ION | | Authors: | Lietzan, A.D, Walton, W.G, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-12-14 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Diverse MarR bacterial regulators of auxin catabolism in the plant microbiome.
Nat Microbiol, 7, 2022
|
|
1BG0
 
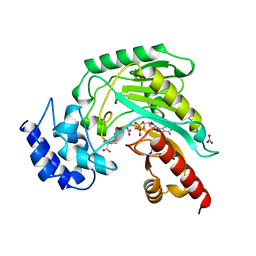 | | TRANSITION STATE STRUCTURE OF ARGININE KINASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ARGININE KINASE, D-ARGININE, ... | | Authors: | Zhou, G, Somasundaram, T, Blanc, E, Parthasarathy, G, Ellington, W.R, Chapman, M.S. | | Deposit date: | 1998-06-03 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Transition state structure of arginine kinase: implications for catalysis of bimolecular reactions.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1Q47
 
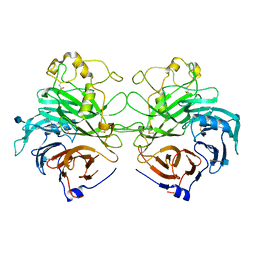 | | Structure of the Semaphorin 3A Receptor-Binding Module | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Semaphorin 3A | | Authors: | Antipenko, A, Himanen, J.-P, van Leyen, K, Nardi-Dei, V, Lesniak, J, Barton, W.A, Rajashankar, K.R, Lu, M, Hoemme, C, Puschel, A, Nikolov, D. | | Deposit date: | 2003-08-01 | | Release date: | 2004-08-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the semaphorin-3A receptor binding module.
Neuron, 39, 2003
|
|
1WY4
 
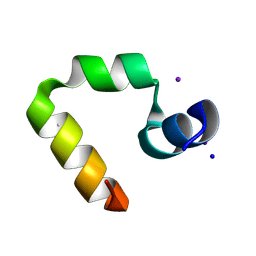 | | Chicken villin subdomain HP-35, K65(NLE), N68H, pH5.1 | | Descriptor: | IODIDE ION, SODIUM ION, Villin | | Authors: | Chiu, T.K, Kubelka, J, Herbst-Irmer, R, Eaton, W.A, Hofrichter, J, Davies, D.R. | | Deposit date: | 2005-02-04 | | Release date: | 2005-05-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | High-resolution x-ray crystal structures of the villin headpiece subdomain, an ultrafast folding protein.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1WY3
 
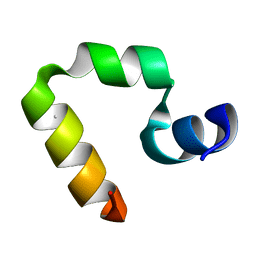 | | Chicken villin subdomain HP-35, K65(NLE), N68H, pH7.0 | | Descriptor: | Villin | | Authors: | Chiu, T.K, Kubelka, J, Herbst-Irmer, R, Eaton, W.A, Hofrichter, J, Davies, D.R. | | Deposit date: | 2005-02-04 | | Release date: | 2005-05-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | High-resolution x-ray crystal structures of the villin headpiece subdomain, an ultrafast folding protein.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
4JYO
 
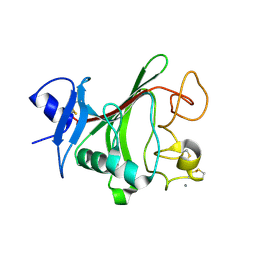 | | Structural basis for angiopoietin-1 mediated signaling initiation | | Descriptor: | Angiopoietin-1, CALCIUM ION | | Authors: | Yu, X, Seegar, T.C.M, Dalton, A.C, Tzvetkova-Robev, D, Goldgur, Y, Nikolov, D.B, Barton, W.A. | | Deposit date: | 2013-03-31 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for angiopoietin-1-mediated signaling initiation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
