2AOX
 
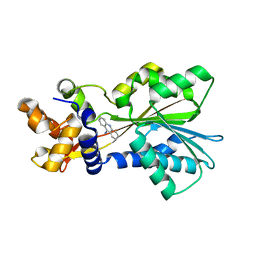 | | Histamine Methyltransferase (Primary Variant T105) Complexed with the Acetylcholinesterase Inhibitor and Altzheimer's Disease Drug Tacrine | | Descriptor: | Histamine N-methyltransferase, TACRINE | | Authors: | Horton, J.R, Sawada, K, Nishibori, M, Cheng, X. | | Deposit date: | 2005-08-14 | | Release date: | 2005-09-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Structural basis for inhibition of histamine N-methyltransferase by diverse drugs
J.Mol.Biol., 353, 2005
|
|
2AOV
 
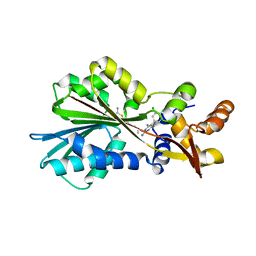 | | Histamine Methyltransferase Complexed with the Antifolate Drug Metoprine | | Descriptor: | 4-(DIMETHYLAMINO)BUTYL IMIDOTHIOCARBAMATE, 5-(3,4-DICHLOROPHENYL)-6-METHYLPYRIMIDINE-2,4-DIAMINE, Histamine N-methyltransferase | | Authors: | Horton, J.R, Sawada, K, Nishibori, M, Cheng, X. | | Deposit date: | 2005-08-14 | | Release date: | 2005-09-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural basis for inhibition of histamine N-methyltransferase by diverse drugs
J.Mol.Biol., 353, 2005
|
|
2CHY
 
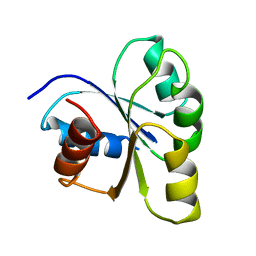 | | THREE-DIMENSIONAL STRUCTURE OF CHEY, THE RESPONSE REGULATOR OF BACTERIAL CHEMOTAXIS | | Descriptor: | CHEY | | Authors: | Mottonen, J.M, Stock, A.M, Stock, J.B, Schutt, C.E. | | Deposit date: | 1990-05-17 | | Release date: | 1990-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Three-dimensional structure of CheY, the response regulator of bacterial chemotaxis.
Nature, 337, 1989
|
|
2ECH
 
 | |
1JOU
 
 | |
2AOU
 
 | | Histamine Methyltransferase Complexed with the Antimalarial Drug Amodiaquine | | Descriptor: | 4-[(7-CHLOROQUINOLIN-4-YL)AMINO]-2-[(DIETHYLAMINO)METHYL]PHENOL, Histamine N-methyltransferase | | Authors: | Horton, J.R, Sawada, K, Nishibori, M, Cheng, X. | | Deposit date: | 2005-08-14 | | Release date: | 2005-09-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for inhibition of histamine N-methyltransferase by diverse drugs
J.Mol.Biol., 353, 2005
|
|
1LQ8
 
 | | Crystal structure of cleaved protein C inhibitor | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Huntington, J.A, Kjellberg, M, Stenflo, J. | | Deposit date: | 2002-05-09 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Protein C Inhibitor Provides Insights into Hormone Binding and Heparin Activation
Structure, 11, 2003
|
|
3I0M
 
 | | Structure of the S. pombe Nbs1 FHA/BRCT-repeat domain | | Descriptor: | DNA repair and telomere maintenance protein nbs1, GLYCEROL | | Authors: | Clapperton, J.A, Lloyd, J, Chapman, J.R, Jackson, S.P, Smerdon, S.J. | | Deposit date: | 2009-06-25 | | Release date: | 2009-10-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A supramodular FHA/BRCT-repeat architecture mediates Nbs1 adaptor function in response to DNA damage
Cell(Cambridge,Mass.), 139, 2009
|
|
2AOT
 
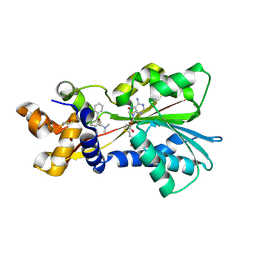 | | Histamine Methyltransferase Complexed with the Antihistamine Drug Diphenhydramine | | Descriptor: | Histamine N-methyltransferase, N-[2-(BENZHYDRYLOXY)ETHYL]-N,N-DIMETHYLAMINE, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Horton, J.R, Sawada, K, Nishibori, M, Cheng, X. | | Deposit date: | 2005-08-14 | | Release date: | 2005-09-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for inhibition of histamine N-methyltransferase by diverse drugs
J.Mol.Biol., 353, 2005
|
|
2AOW
 
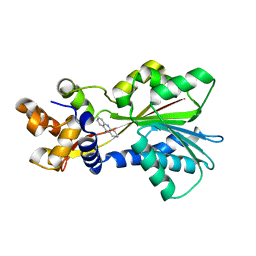 | | Histamine Methyltransferase (Natural Variant I105) Complexed with the Acetylcholinesterase Inhibitor and Altzheimer's Disease Drug Tacrine | | Descriptor: | Histamine N-methyltransferase, TACRINE | | Authors: | Horton, J.R, Sawada, K, Nishibori, M, Cheng, X. | | Deposit date: | 2005-08-14 | | Release date: | 2005-09-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Structural basis for inhibition of histamine N-methyltransferase by diverse drugs
J.Mol.Biol., 353, 2005
|
|
2FKC
 
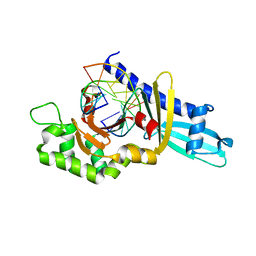 | |
2FLC
 
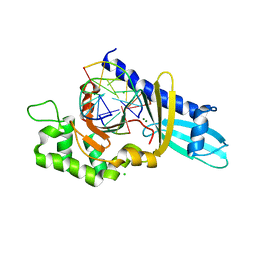 | |
2FL3
 
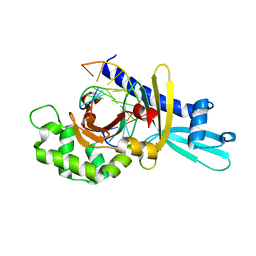 | | Binary Complex of Restriction Endonuclease HinP1I with Cognate DNA | | Descriptor: | 5'-D(*CP*CP*AP*GP*CP*GP*CP*TP*GP*G)-3', R.HinP1I Restriction Endonuclease | | Authors: | Horton, J.R. | | Deposit date: | 2006-01-05 | | Release date: | 2006-02-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | DNA nicking by HinP1I endonuclease: bending, base flipping and minor groove expansion.
Nucleic Acids Res., 34, 2006
|
|
3IG0
 
 | |
3EVJ
 
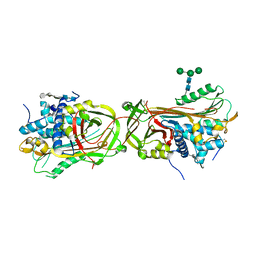 | | Intermediate structure of antithrombin bound to the natural pentasaccharide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-3,6-di-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-methyl 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranoside, ... | | Authors: | Huntington, J.A, Belzar, K.J. | | Deposit date: | 2008-10-13 | | Release date: | 2008-10-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The critical role of hinge-region expulsion in the induced-fit heparin binding mechanism of antithrombin.
J. Mol. Biol., 386, 2009
|
|
1RB5
 
 | |
3ICV
 
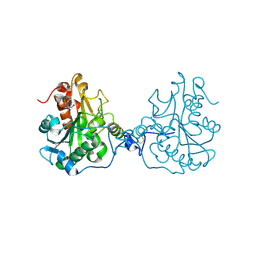 | | Structural Consequences of a Circular Permutation on Lipase B from Candida Antartica | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Lipase B | | Authors: | Horton, J.R, Qian, Z, Jia, D, Lutz, S, Cheng, X. | | Deposit date: | 2009-07-18 | | Release date: | 2009-10-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural redesign of lipase B from Candida antarctica by circular permutation and incremental truncation.
J.Mol.Biol., 393, 2009
|
|
3HSS
 
 | | A higher resolution structure of Rv0554 from Mycobacterium tuberculosis complexed with malonic acid | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Johnston, J.M, Baker, E.N. | | Deposit date: | 2009-06-10 | | Release date: | 2010-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional analysis of Rv0554 from Mycobacterium tuberculosis: testing a putative role in menaquinone biosynthesis.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3HYS
 
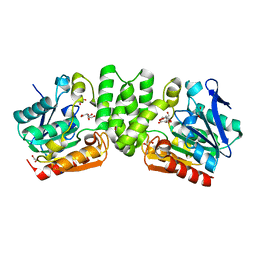 | | Structure of Rv0554 from Mycobacterium tuberculosis complexed with Malonic Acid | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MALONIC ACID, ... | | Authors: | Johnston, J.M, Baker, E.N. | | Deposit date: | 2009-06-23 | | Release date: | 2010-06-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional analysis of Rv0554 from Mycobacterium tuberculosis: testing a putative role in menaquinone biosynthesis.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3HZO
 
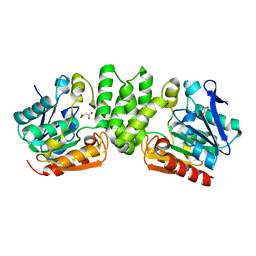 | |
3ICW
 
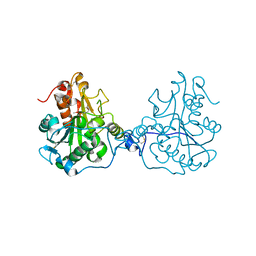 | | Structure of a Circular Permutation on Lipase B from Candida Antartica with Bound Suicide Inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lipase B, PHOSPHATE ION, ... | | Authors: | Horton, J.R, Qian, Z, Jia, D, Lutz, S.A, Cheng, X. | | Deposit date: | 2009-07-18 | | Release date: | 2009-10-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural redesign of lipase B from Candida antarctica by circular permutation and incremental truncation.
J.Mol.Biol., 393, 2009
|
|
3IFZ
 
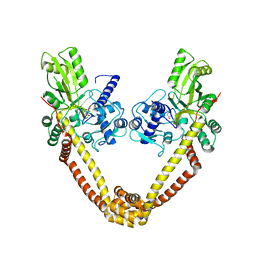 | | crystal structure of the first part of the Mycobacterium tuberculosis DNA gyrase reaction core: the breakage and reunion domain at 2.7 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA gyrase subunit A, SODIUM ION | | Authors: | Piton, J, Aubry, A, Delarue, M, Mayer, C. | | Deposit date: | 2009-07-27 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the quinolone resistance mechanism of Mycobacterium tuberculosis DNA gyrase.
Plos One, 5, 2010
|
|
2G1P
 
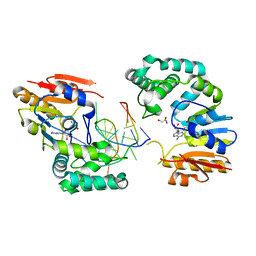 | | Structure of E. coli DNA adenine methyltransferase (DAM) | | Descriptor: | 5'-D(*TP*CP*TP*AP*GP*AP*TP*CP*TP*AP*GP*A)-3', DNA adenine methylase, GLYCEROL, ... | | Authors: | Horton, J.R, Liebert, K, Bekes, M, Jeltsch, A, Cheng, X. | | Deposit date: | 2006-02-14 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure and substrate recognition of the Escherichia coli DNA adenine methyltransferase.
J.Mol.Biol., 358, 2006
|
|
1RB4
 
 | |
1TM9
 
 | | NMR Structure of gene target number gi3844938 from Mycoplasma genitalium: Berkeley Structural Genomics Center | | Descriptor: | Hypothetical protein MG354 | | Authors: | Pelton, J.G, Shi, J, Yokota, H, Kim, R, Wemmer, D.E, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-06-10 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Gene Target gi3844938 from Mycoplasma genitalium
To be Published
|
|
