1YYJ
 
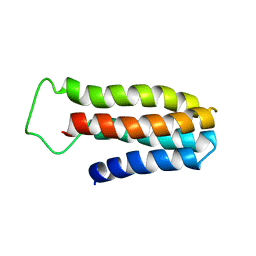 | | The NMR solution structure of a redesigned apocytochrome b562:Rd-apocyt b562 | | 分子名称: | redesigned apocytochrome B562 | | 著者 | Feng, H, Takei, J, Lipsitz, R, Tjandra, N, Bai, Y, Berkeley Structural Genomics Center (BSGC) | | 登録日 | 2005-02-25 | | 公開日 | 2005-08-25 | | 最終更新日 | 2023-09-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Specific non-native hydrophobic interactions in a hidden folding intermediate: implications for protein folding
Biochemistry, 42, 2003
|
|
6UD0
 
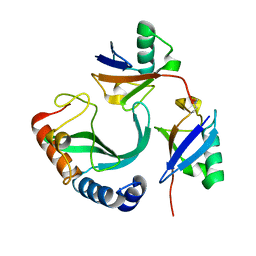 | | Solution-state NMR structural ensemble of human Tsg101 UEV in complex with K63-linked diubiquitin | | 分子名称: | Tumor susceptibility gene 101 protein, Ubiquitin | | 著者 | Strickland, M, Watanabe, S, Bonn, S.M, Camara, C.M, Fushman, D, Carter, C.A, Tjandra, N. | | 登録日 | 2019-09-18 | | 公開日 | 2021-03-17 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Tsg101/ESCRT-I Recruitment Regulated by the Dual Binding Modes of K63-Linked Diubiquitin
Structure, 2021
|
|
8V3N
 
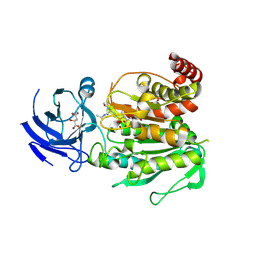 | | CCP5 in complex with Glu-P-Glu transition state analog | | 分子名称: | (2S)-2-{[(S)-[(3S)-3-acetamido-4-(ethylamino)-4-oxobutyl](hydroxy)phosphoryl]methyl}pentanedioic acid, Cytosolic carboxypeptidase-like protein 5, D-MALATE, ... | | 著者 | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | 登録日 | 2023-11-28 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
8V3S
 
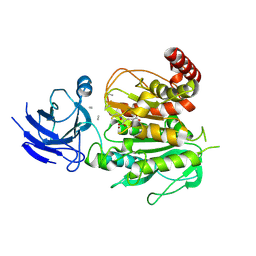 | | Structure of CCP5 class3 | | 分子名称: | Cytosolic carboxypeptidase-like protein 5, GLUTAMIC ACID, ZINC ION, ... | | 著者 | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | 登録日 | 2023-11-28 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-08-07 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
8V4K
 
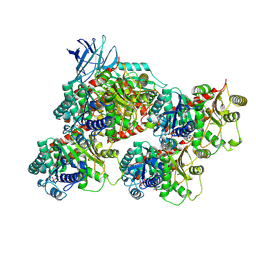 | | CCP5 in complex with microtubules class1 | | 分子名称: | Cytosolic carboxypeptidase-like protein 5, GLUTAMIC ACID, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | 登録日 | 2023-11-29 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-08-07 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
8V3O
 
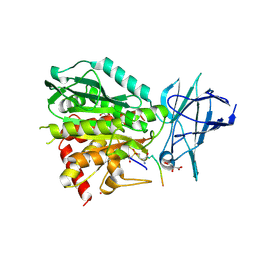 | | CCP5 in complex with Glu-P-peptide 1 transition state analog | | 分子名称: | Cytosolic carboxypeptidase-like protein 5, D-MALATE, POTASSIUM ION, ... | | 著者 | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | 登録日 | 2023-11-28 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
8V4M
 
 | | CCP5 in complex with microtubules class3 | | 分子名称: | Cytosolic carboxypeptidase-like protein 5, GLUTAMIC ACID, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | 登録日 | 2023-11-29 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-08-07 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
8V3M
 
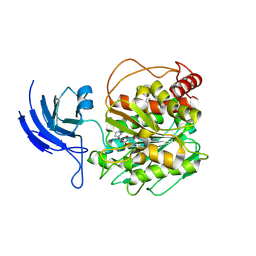 | | CCP5 apo structure | | 分子名称: | Cytosolic carboxypeptidase-like protein 5, D-MALATE, IMIDAZOLE, ... | | 著者 | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | 登録日 | 2023-11-28 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
8V3R
 
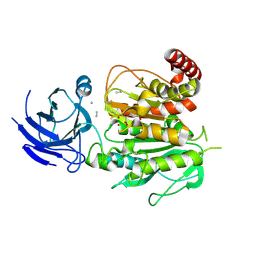 | | Structure of CCP5 class2 | | 分子名称: | Cytosolic carboxypeptidase-like protein 5, GLUTAMIC ACID, ZINC ION, ... | | 著者 | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | 登録日 | 2023-11-28 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-08-07 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
8V3P
 
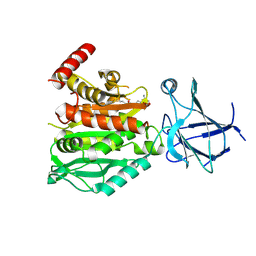 | | CCP5 in complex with Glu-P-peptide 2 transition state analog | | 分子名称: | Cytosolic carboxypeptidase-like protein 5, Tubulin beta-2A chain, ZINC ION | | 著者 | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | 登録日 | 2023-11-28 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (2.36 Å) | | 主引用文献 | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
8V3Q
 
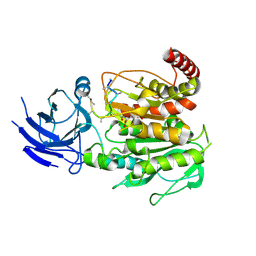 | | Structure of CCP5 class1 | | 分子名称: | Cytosolic carboxypeptidase-like protein 5, GLUTAMIC ACID, ZINC ION, ... | | 著者 | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | 登録日 | 2023-11-28 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-08-07 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
8V4L
 
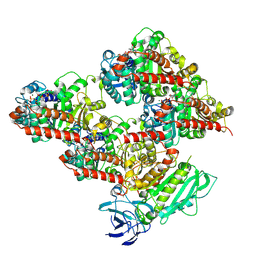 | | CCP5 in complex with microtubules class2 | | 分子名称: | Cytosolic carboxypeptidase-like protein 5, GLUTAMIC ACID, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | 登録日 | 2023-11-29 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-08-07 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
5T1O
 
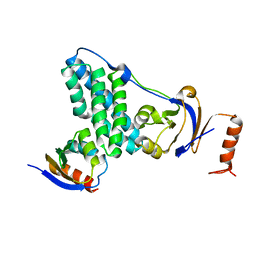 | | Solution-state NMR and SAXS structural ensemble of NPr (1-85) in complex with EIN-Ntr (170-424) | | 分子名称: | Phosphocarrier protein NPr, Phosphoenolpyruvate-protein phosphotransferase PtsP | | 著者 | Strickland, M, Stanley, A.M, Wang, G, Schwieters, C.D, Buchanan, S, Peterkofsky, A, Tjandra, N. | | 登録日 | 2016-08-19 | | 公開日 | 2016-11-16 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR, SOLUTION SCATTERING | | 主引用文献 | Structure of the NPr:EIN(Ntr) Complex: Mechanism for Specificity in Paralogous Phosphotransferase Systems.
Structure, 24, 2016
|
|
5T1N
 
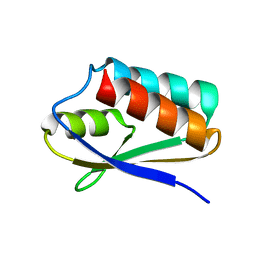 | |
6VZQ
 
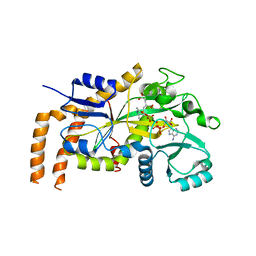 | | Engineered TTLL6 mutant bound to alpha-elongation analog | | 分子名称: | (2~{S})-2-[[[(1~{R})-1-acetamido-4-oxidanyl-4-oxidanylidene-butyl]-phosphonooxy-phosphoryl]methyl]pentanedioic acid, (2~{S})-2-[[[(1~{S})-1-acetamidoethyl]-phosphonooxy-phosphoryl]methyl]pentanedioic acid, ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | Mahalingan, K.K, Keenen, E.K, Strickland, E.K, Li, Y, Liu, Y, Ball, H.L, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | 登録日 | 2020-02-28 | | 公開日 | 2020-08-12 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.08 Å) | | 主引用文献 | Structural basis for polyglutamate chain initiation and elongation by TTLL family enzymes.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VZT
 
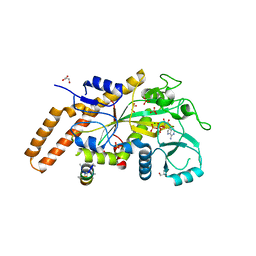 | | TTLL6 bound to ATP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Mahalingan, K.K, Keenen, E.K, Strickland, M, Li, Y, Liu, Y, Ball, H.L, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | 登録日 | 2020-02-28 | | 公開日 | 2020-08-12 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Structural basis for polyglutamate chain initiation and elongation by TTLL family enzymes.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VZV
 
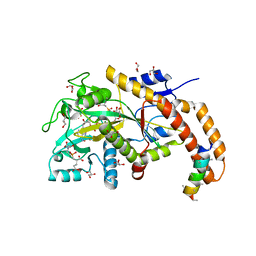 | | TTLL6 bound to gamma-elongation analog | | 分子名称: | (2~{S})-2-[[[(3~{R})-3-acetamido-4-oxidanyl-4-oxidanylidene-butyl]-phosphonooxy-phosphoryl]methyl]pentanedioic acid, (2~{S})-2-[[[(3~{S})-3-acetamido-4-oxidanyl-4-oxidanylidene-butyl]-oxidanyl-phosphoryl]methyl]pentanedioic acid, ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | Mahalingan, K.K, Keenen, E.K, Strickland, M, Li, Y, Liu, Y, Ball, H.L, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | 登録日 | 2020-02-28 | | 公開日 | 2020-08-12 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.33 Å) | | 主引用文献 | Structural basis for polyglutamate chain initiation and elongation by TTLL family enzymes.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VZR
 
 | | Engineered TTLL6 bound to the initiation analog | | 分子名称: | (2~{S})-2-[[[(3~{R})-3-acetamido-4-(ethylamino)-4-oxidanylidene-butyl]-phosphonooxy-phosphoryl]methyl]pentanedioic acid, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | 著者 | Mahalingan, K.K, Keenen, E.K, Strickland, M, Li, Y, Liu, Y, Ball, H.L, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | 登録日 | 2020-02-28 | | 公開日 | 2020-08-12 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural basis for polyglutamate chain initiation and elongation by TTLL family enzymes.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VZU
 
 | | TTLL6 bound to alpha-elongation analog | | 分子名称: | (2~{S})-2-[[[(1~{R})-1-acetamido-4-oxidanyl-4-oxidanylidene-butyl]-oxidanyl-phosphoryl]methyl]pentanedioic acid, (2~{S})-2-[[[(1~{R})-1-acetamido-4-oxidanyl-4-oxidanylidene-butyl]-phosphonooxy-phosphoryl]methyl]pentanedioic acid, ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | Mahalingan, K.K, Keenen, E.K, Strickland, M, Li, Y, Liu, Y, Ball, H.B, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | 登録日 | 2020-02-28 | | 公開日 | 2020-08-12 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Structural basis for polyglutamate chain initiation and elongation by TTLL family enzymes.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VZS
 
 | | Engineered TTLL6 mutant bound to gamma-elongation analog | | 分子名称: | (2~{S})-2-[[[(3~{R})-3-acetamido-4-oxidanyl-4-oxidanylidene-butyl]-phosphonooxy-phosphoryl]methyl]pentanedioic acid, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | 著者 | Mahalingan, K.K, Keenen, E.K, Strickland, M, Li, Y, Liu, Y, Ball, H.L, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | 登録日 | 2020-02-28 | | 公開日 | 2020-08-12 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.66 Å) | | 主引用文献 | Structural basis for polyglutamate chain initiation and elongation by TTLL family enzymes.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VZW
 
 | | TTLL6 bound to the initiation analog | | 分子名称: | (2~{S})-2-[[[(3~{R})-3-acetamido-4-(ethylamino)-4-oxidanylidene-butyl]-phosphonooxy-phosphoryl]methyl]pentanedioic acid, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | 著者 | Mahalingan, K.K, Keenen, E.K, Strickland, M, Li, Y, Liu, Y, Ball, H.L, Tanner, T.E, Tjandra, N, Roll-Mecak, A. | | 登録日 | 2020-02-28 | | 公開日 | 2020-08-12 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural basis for polyglutamate chain initiation and elongation by TTLL family enzymes.
Nat.Struct.Mol.Biol., 27, 2020
|
|
2JZ7
 
 | |
2EZA
 
 | | AMINO TERMINAL DOMAIN OF ENZYME I FROM ESCHERICHIA COLI, NMR, RESTRAINED REGULARIZED MEAN STRUCTURE | | 分子名称: | PHOSPHOTRANSFERASE SYSTEM, ENZYME I | | 著者 | Clore, G.M, Tjandra, N, Garrett, D.S, Gronenborn, A.M. | | 登録日 | 1997-05-07 | | 公開日 | 1997-08-20 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Defining long range order in NMR structure determination from the dependence of heteronuclear relaxation times on rotational diffusion anisotropy.
Nat.Struct.Biol., 4, 1997
|
|
2EZC
 
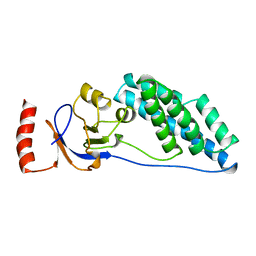 | | AMINO TERMINAL DOMAIN OF ENZYME I FROM ESCHERICHIA COLI, NMR, 14 STRUCTURES | | 分子名称: | PHOSPHOTRANSFERASE SYSTEM, ENZYME I | | 著者 | Clore, G.M, Tjandra, N, Garrett, D.S, Gronenborn, A.M. | | 登録日 | 1997-05-07 | | 公開日 | 1997-08-20 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Defining long range order in NMR structure determination from the dependence of heteronuclear relaxation times on rotational diffusion anisotropy.
Nat.Struct.Biol., 4, 1997
|
|
2EZB
 
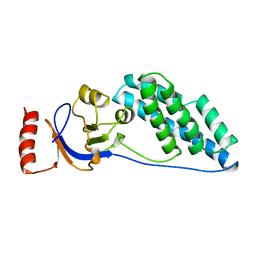 | | AMINO TERMINAL DOMAIN OF ENZYME I FROM ESCHERICHIA COLI, NMR, 14 STRUCTURES | | 分子名称: | PHOSPHOTRANSFERASE SYSTEM, ENZYME I | | 著者 | Clore, G.M, Tjandra, N, Garrett, D.S, Gronenborn, A.M. | | 登録日 | 1997-05-07 | | 公開日 | 1997-08-20 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Defining long range order in NMR structure determination from the dependence of heteronuclear relaxation times on rotational diffusion anisotropy.
Nat.Struct.Biol., 4, 1997
|
|
