7LXE
 
 | |
6RTZ
 
 | |
6RU0
 
 | |
1KDD
 
 | | X-ray structure of the coiled coil GCN4 ACID BASE HETERODIMER ACID-d12La16I BASE-d12La16L | | Descriptor: | GCN4 ACID BASE HETERODIMER ACID-d12La16I, GCN4 ACID BASE HETERODIMER BASE-d12La16L | | Authors: | Keating, A.E, Malashkevich, V.N, Tidor, B, Kim, P.S. | | Deposit date: | 2001-11-12 | | Release date: | 2001-11-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Side-chain repacking calculations for predicting structures and stabilities of heterodimeric coiled coils.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1KD8
 
 | | X-RAY STRUCTURE OF THE COILED COIL GCN4 ACID BASE HETERODIMER ACID-d12Ia16V BASE-d12La16L | | Descriptor: | GCN4 ACID BASE HETERODIMER ACID-d12Ia16V, GCN4 ACID BASE HETERODIMER BASE-d12La16L | | Authors: | Keating, A.E, Malashkevich, V.N, Tidor, B, Kim, P.S. | | Deposit date: | 2001-11-12 | | Release date: | 2001-11-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Side-chain repacking calculations for predicting structures and stabilities of heterodimeric coiled coils.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1KD9
 
 | | X-RAY STRUCTURE OF THE COILED COIL GCN4 ACID BASE HETERODIMER ACID-d12La16L BASE-d12La16L | | Descriptor: | GCN4 ACID BASE HETERODIMER ACID-d12La16L, GCN4 ACID BASE HETERODIMER BASE-d12La16L | | Authors: | Keating, A.E, Malashkevich, V.N, Tidor, B, Kim, P.S. | | Deposit date: | 2001-11-12 | | Release date: | 2001-11-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Side-chain repacking calculations for predicting structures and stabilities of heterodimeric coiled coils.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
2V4B
 
 | | Crystal Structure of Human ADAMTS-1 catalytic Domain and Cysteine- Rich Domain (apo-form) | | Descriptor: | ADAMTS-1, CADMIUM ION, MAGNESIUM ION, ... | | Authors: | Gerhardt, S, Hassall, G, Hawtin, P, McCall, E, Flavell, L, Minshull, C, Hargreaves, D, Ting, A, Pauptit, R.A, Parker, A.E, Abbott, W.M. | | Deposit date: | 2007-06-28 | | Release date: | 2008-01-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Human Adamts-1 Reveal a Conserved Catalytic Domain and a Disintegrin-Like Domain with a Fold Homologous to Cysteine-Rich Domains.
J.Mol.Biol., 373, 2007
|
|
2JIH
 
 | | Crystal Structure of Human ADAMTS-1 catalytic Domain and Cysteine- Rich Domain (complex-form) | | Descriptor: | (2S,3R)-N~4~-[(1S)-2,2-dimethyl-1-(methylcarbamoyl)propyl]-N~1~,2-dihydroxy-3-(2-methylpropyl)butanediamide, ADAMTS-1, CADMIUM ION, ... | | Authors: | Gerhardt, S, Hassall, G, Hawtin, P, McCall, E, Flavell, L, Minshull, C, Hargreaves, D, Ting, A, Pauptit, R.A, Parker, A.E, Abbott, W.M. | | Deposit date: | 2007-06-28 | | Release date: | 2008-01-15 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of human ADAMTS-1 reveal a conserved catalytic domain and a disintegrin-like domain with a fold homologous to cysteine-rich domains.
J. Mol. Biol., 373, 2007
|
|
6T3B
 
 | | Crystal structure of PI3Kgamma with a dihydropurinone inhibitor (compound 4) | | Descriptor: | 2-[(4-methoxy-2-methyl-phenyl)amino]-7-methyl-9-(4-oxidanylcyclohexyl)purin-8-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Petersen, J, Oster, L, Schimpl, M, Goldberg, F.W, Finlay, M.R.V, Ting, A.K.T, Beattie, D, Lamont, G.M, Fallan, C, Wrigley, G.L, Howard, M.R, Williamson, B, Davies, B.R, Cadogan, E.B, Ramos-Montoya, A, Dean, E. | | Deposit date: | 2019-10-10 | | Release date: | 2020-01-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | The Discovery of 7-Methyl-2-[(7-methyl[1,2,4]triazolo[1,5-a]pyridin-6-yl)amino]-9-(tetrahydro-2H-pyran-4-yl)-7,9-dihydro-8H-purin-8-one (AZD7648), a Potent and Selective DNA-Dependent Protein Kinase (DNA-PK) Inhibitor.
J.Med.Chem., 63, 2020
|
|
6T3C
 
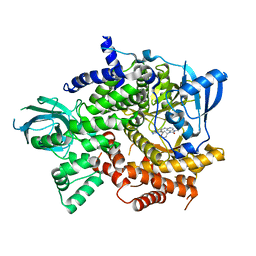 | | Crystal structure of PI3Kgamma in complex with DNA-PK inhibitor AZD7648 | | Descriptor: | 7-methyl-2-[(7-methyl-[1,2,4]triazolo[1,5-a]pyridin-6-yl)amino]-9-(oxan-4-yl)purin-8-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Schimpl, M, Goldberg, F.W, Finlay, M.R.V, Ting, A.K.T, Beattie, D, Lamont, G.M, Fallan, C, Wrigley, G.L, Howard, M.R, Williamson, B, Davies, B.R, Cadogan, E.B, Ramos-Montoya, A, Dean, E. | | Deposit date: | 2019-10-10 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | The Discovery of 7-Methyl-2-[(7-methyl[1,2,4]triazolo[1,5-a]pyridin-6-yl)amino]-9-(tetrahydro-2H-pyran-4-yl)-7,9-dihydro-8H-purin-8-one (AZD7648), a Potent and Selective DNA-Dependent Protein Kinase (DNA-PK) Inhibitor.
J.Med.Chem., 63, 2020
|
|
6T2W
 
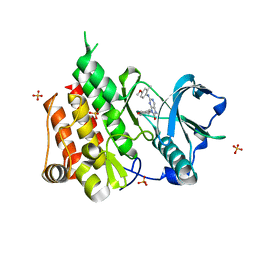 | | Crystal structure of the CSF1R kinase domain with a dihydropurinone inhibitor (compound 4) | | Descriptor: | 2-[(4-methoxy-2-methyl-phenyl)amino]-7-methyl-9-(4-oxidanylcyclohexyl)purin-8-one, Macrophage colony-stimulating factor 1 receptor, SULFATE ION | | Authors: | Schimpl, M, Goldberg, F.W, Finlay, M.R.V, Ting, A.K.T, Beattie, D, Lamont, G.M, Fallan, C, Wrigley, G.L, Howard, M.R, Williamson, B, Davies, B.R, Cadogan, E.B, Ramos-Montoya, A, Dean, E. | | Deposit date: | 2019-10-09 | | Release date: | 2020-01-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Discovery of 7-Methyl-2-[(7-methyl[1,2,4]triazolo[1,5-a]pyridin-6-yl)amino]-9-(tetrahydro-2H-pyran-4-yl)-7,9-dihydro-8H-purin-8-one (AZD7648), a Potent and Selective DNA-Dependent Protein Kinase (DNA-PK) Inhibitor.
J.Med.Chem., 63, 2020
|
|
7Z61
 
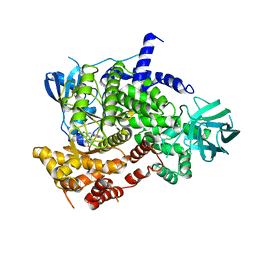 | | Crystal structure of PI3Kgamma with a dihydropurinone inhibitor (compound 18) | | Descriptor: | 9-[(3~{R},4~{R})-4-fluoranylpyrrolidin-3-yl]-7-methyl-2-[(7-methylquinolin-6-yl)amino]purin-8-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Goldberg, F.W, Ting, A.K.T, Schimpl, M. | | Deposit date: | 2022-03-10 | | Release date: | 2022-07-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.738 Å) | | Cite: | Optimization of hERG and Pharmacokinetic Properties for Basic Dihydro-8 H -purin-8-one Inhibitors of DNA-PK.
Acs Med.Chem.Lett., 13, 2022
|
|
6MBB
 
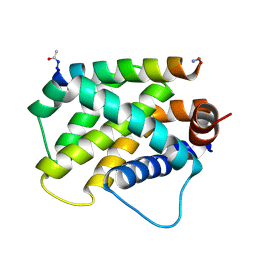 | | Human Bfl-1 in complex with the designed peptide dF1 | | Descriptor: | Bcl-2-related protein A1, dF1 | | Authors: | Jenson, J.M, Keating, A.E. | | Deposit date: | 2018-08-29 | | Release date: | 2019-03-06 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Tertiary Structural Motif Sequence Statistics Enable Facile Prediction and Design of Peptides that Bind Anti-apoptotic Bfl-1 and Mcl-1.
Structure, 27, 2019
|
|
2PQK
 
 | | X-ray crystal structure of human Mcl-1 in complex with Bim BH3 | | Descriptor: | Bim BH3 peptide, Induced myeloid leukemia cell differentiation protein Mcl-1, SODIUM ION, ... | | Authors: | Bare, E, Grant, R.A, Keating, A.E. | | Deposit date: | 2007-05-02 | | Release date: | 2007-06-19 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mcl-1-Bim complexes accommodate surprising point mutations via minor structural changes.
Protein Sci., 19, 2010
|
|
5P21
 
 | |
3BH6
 
 | | Crystal structure of the RP2-Arl3 complex bound to GppNHp | | Descriptor: | ADP-ribosylation factor-like protein 3, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Veltel, S, Gasper, R, Wittinghofer, A. | | Deposit date: | 2007-11-28 | | Release date: | 2008-03-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The retinitis pigmentosa 2 gene product is a GTPase-activating protein for Arf-like 3
Nat.Struct.Mol.Biol., 15, 2008
|
|
7LXF
 
 | |
3KJ0
 
 | | Mcl-1 in complex with Bim BH3 mutant I2dY | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bcl-2-like protein 11, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Fire, E, Grant, R.A, Keating, A.E. | | Deposit date: | 2009-11-02 | | Release date: | 2010-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mcl-1-Bim complexes accommodate surprising point mutations via minor structural changes.
Protein Sci., 19, 2010
|
|
3KJ2
 
 | | Mcl-1 in complex with Bim BH3 mutant F4aE | | Descriptor: | ACETATE ION, Bcl-2-like protein 11, Induced myeloid leukemia cell differentiation protein Mcl-1, ... | | Authors: | Fire, E, Grant, R.A, Keating, A.E. | | Deposit date: | 2009-11-02 | | Release date: | 2010-02-16 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Mcl-1-Bim complexes accommodate surprising point mutations via minor structural changes.
Protein Sci., 19, 2010
|
|
1HVQ
 
 | | CRYSTAL STRUCTURES OF HEVAMINE, A PLANT DEFENCE PROTEIN WITH CHITINASE AND LYSOZYME ACTIVITY, AND ITS COMPLEX WITH AN INHIBITOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HEVAMINE A | | Authors: | Terwisscha Van Scheltinga, A.C, Kalk, K.H, Beintema, J.J, Dijkstra, B.W. | | Deposit date: | 1994-10-13 | | Release date: | 1995-12-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of hevamine, a plant defence protein with chitinase and lysozyme activity, and its complex with an inhibitor.
Structure, 2, 1994
|
|
221P
 
 | | THREE-DIMENSIONAL STRUCTURES OF H-RAS P21 MUTANTS: MOLECULAR BASIS FOR THEIR INABILITY TO FUNCTION AS SIGNAL SWITCH MOLECULES | | Descriptor: | H-RAS P21 PROTEIN, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Krengel, U, Scherer, A, Kabsch, W, Wittinghofer, A, Pai, E.F. | | Deposit date: | 1991-06-06 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structures of H-ras p21 mutants: molecular basis for their inability to function as signal switch molecules.
Cell(Cambridge,Mass.), 62, 1990
|
|
1GNR
 
 | | X-RAY CRYSTAL STRUCTURE ANALYSIS OF THE CATALYTIC DOMAIN OF THE ONCOGENE PRODUCT P21H-RAS COMPLEXED WITH CAGED GTP AND MANT DGPPNHP | | Descriptor: | C-H-RAS P21 PROTEIN, GUANOSINE 5'-TRIPHOSPHATE P3-[1-(2-NITROPHENYL)ETHYL ESTER], MAGNESIUM ION | | Authors: | Scheidig, A, Franken, S.M, Corrie, J.E.T, Reid, G.P, Wittinghofer, A, Pai, E.F, Goody, R.S. | | Deposit date: | 1995-05-11 | | Release date: | 1995-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray crystal structure analysis of the catalytic domain of the oncogene product p21H-ras complexed with caged GTP and mant dGppNHp.
J.Mol.Biol., 253, 1995
|
|
3BRW
 
 | | Structure of the Rap-RapGAP complex | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Scrima, A, Thomas, C, Deaconescu, D, Wittinghofer, A. | | Deposit date: | 2007-12-21 | | Release date: | 2008-03-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The Rap-RapGAP complex: GTP hydrolysis without catalytic glutamine and arginine residues
Embo J., 27, 2008
|
|
6UA3
 
 | |
3KJ1
 
 | | Mcl-1 in complex with Bim BH3 mutant I2dA | | Descriptor: | ACETATE ION, Bcl-2-like protein 11, CHLORIDE ION, ... | | Authors: | Fire, E, Grant, R.A, Keating, A.E. | | Deposit date: | 2009-11-02 | | Release date: | 2010-02-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.945 Å) | | Cite: | Mcl-1-Bim complexes accommodate surprising point mutations via minor structural changes.
Protein Sci., 19, 2010
|
|
