4LPV
 
 | | Crystal structure of TENCON variant P41BR3-42 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, TENCON variant P41BR3-42 | | Authors: | Teplyakov, A, Obmolova, G, Luo, J, Gilliland, G.L. | | Deposit date: | 2013-07-16 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | C-terminal beta-strand swapping in a consensus-derived fibronectin Type III scaffold.
Proteins, 82, 2014
|
|
4LPT
 
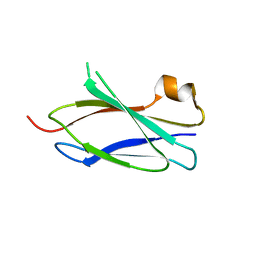 | |
4LPX
 
 | |
1NRK
 
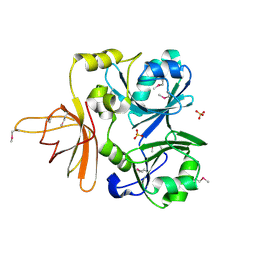 | |
4M6N
 
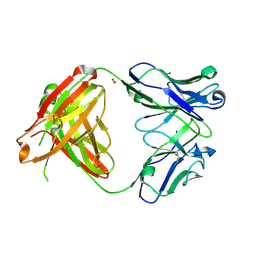 | |
4M7K
 
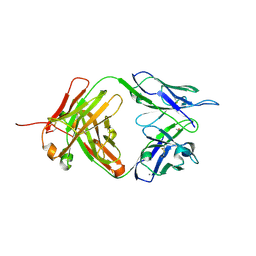 | | Crystal structure of anti-tissue factor antibody 10H10 | | Descriptor: | 10H10 heavy chain, 10H10 light chain, ACETATE ION, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2013-08-12 | | Release date: | 2014-03-26 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antibody modeling assessment II. Structures and models.
Proteins, 82, 2014
|
|
1M68
 
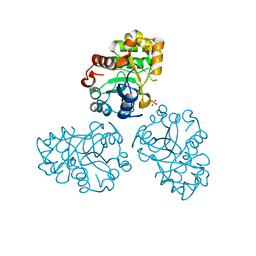 | | YCDX PROTEIN, TRINUCLEAR ZINC SITE | | Descriptor: | Hypothetical protein ycdX, SULFATE ION, ZINC ION | | Authors: | Teplyakov, A, Obmolova, G, Khil, P.P, Camerini-Otero, R.D, Gilliland, G.L. | | Deposit date: | 2002-07-14 | | Release date: | 2003-04-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the Escherichia coli YcdX protein reveals a
trinuclear zinc active site
PROTEINS: STRUCT.,FUNCT.,GENET., 51, 2003
|
|
4M6M
 
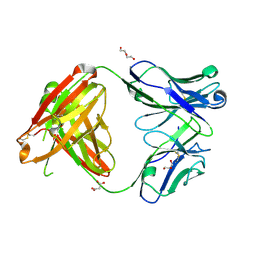 | | Crystal structure of anti-IL-23 antibody CNTO1959 at pH 9.5 | | Descriptor: | CNTO1959 heavy chain, CNTO1959 light chain, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Teplyakov, A, Obmolova, G, Gilliland, G.L. | | Deposit date: | 2013-08-09 | | Release date: | 2014-03-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Antibody modeling assessment II. Structures and models.
Proteins, 82, 2014
|
|
1NOA
 
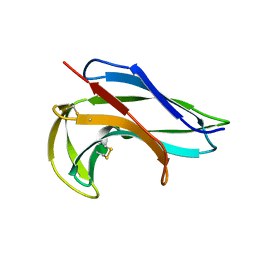 | |
1M65
 
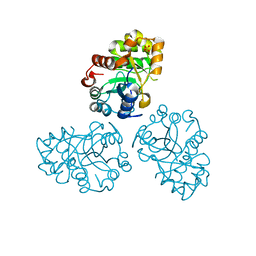 | | YCDX PROTEIN | | Descriptor: | Hypothetical protein ycdX, SODIUM ION, SULFATE ION, ... | | Authors: | Teplyakov, A, Obmolova, G, Khil, P.P, Camerini-Otero, R.D, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2002-07-12 | | Release date: | 2003-04-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal structure of the Escherichia coli YcdX protein reveals a trinuclear zinc active site
PROTEINS: STRUCT.,FUNCT.,GENET., 51, 2003
|
|
1FL9
 
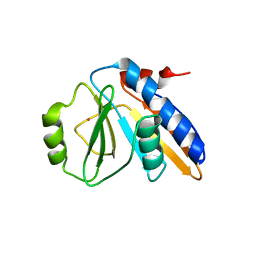 | | THE YJEE PROTEIN | | Descriptor: | HYPOTHETICAL PROTEIN HI0065, MERCURY (II) ION | | Authors: | Teplyakov, A, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2000-08-13 | | Release date: | 2002-08-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the YjeE protein from Haemophilus influenzae: a putative Atpase involved in cell wall synthesis
Proteins, 48, 2002
|
|
4M6O
 
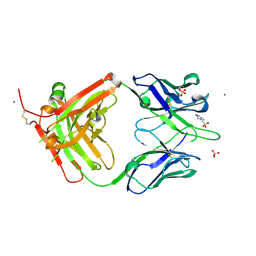 | | Crystal structure of anti-NGF antibody CNTO7309 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CNTO7309 heavy chain, CNTO7309 light chain, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2013-08-09 | | Release date: | 2014-03-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Antibody modeling assessment II. Structures and models.
Proteins, 82, 2014
|
|
4MAU
 
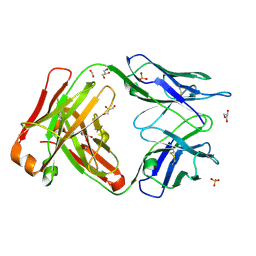 | | Crystal structure of anti-ST2L antibody C2244 | | Descriptor: | C2244 heavy chain, C2244 light chain, FORMIC ACID, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2013-08-16 | | Release date: | 2014-03-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antibody modeling assessment II. Structures and models.
Proteins, 82, 2014
|
|
4PS4
 
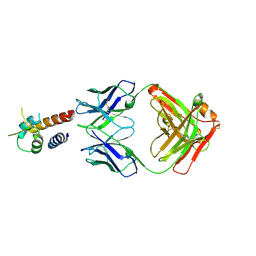 | | Crystal structure of the complex between IL-13 and M1295 FAB | | Descriptor: | Interleukin-13, M1295 HEAVY CHAIN, M1295 LIGHT CHAIN | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2014-03-06 | | Release date: | 2014-03-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Human Framework Adaptation of a Mouse Anti-Human Il-13 Antibody.
J.Mol.Biol., 398, 2010
|
|
6XLI
 
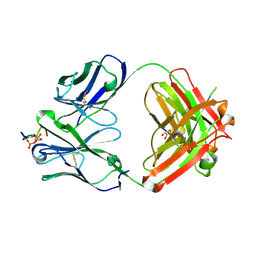 | | CRYSTAL STRUCTURE OF ANTI-TAU ANTIBODY PT3 Fab+pT212/pT217-TAU PEPTIDE | | Descriptor: | GLYCEROL, PT3 Fab Heavy Chain, PT3 Fab Light Chain, ... | | Authors: | Malia, T.J, Teplyakov, A, Luo, J. | | Deposit date: | 2020-06-28 | | Release date: | 2020-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and Functional Characterization of hPT3, a Humanized Anti-Phospho Tau Selective Monoclonal Antibody.
J Alzheimers Dis, 77, 2020
|
|
1LQA
 
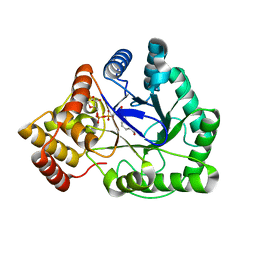 | | TAS PROTEIN FROM ESCHERICHIA COLI IN COMPLEX WITH NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Tas protein | | Authors: | Obmolova, G, Teplyakov, A, Khil, P.P, Howard, A.J, Camerini-Otero, R.D, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2002-05-09 | | Release date: | 2003-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | Crystal structure of the Escherichia coli Tas protein, an NADP(H)-dependent aldo-keto reductase
PROTEINS: STRUCT.,FUNCT.,GENET., 53, 2003
|
|
4DN3
 
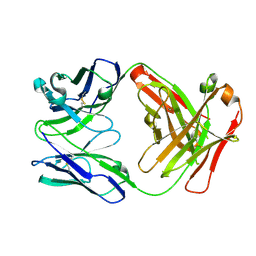 | | Crystal structure of anti-mcp-1 antibody cnto888 | | Descriptor: | CNTO888 HEAVY CHAIN, CNTO888 LIGHT CHAIN | | Authors: | Obmolova, G, Teplyakov, A, Malia, T, Grygiel, T, Sweet, R, Snyder, L, Gilliland, G. | | Deposit date: | 2012-02-08 | | Release date: | 2012-10-03 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for high selectivity of anti-CCL2 neutralizing antibody CNTO 888.
Mol.Immunol., 51, 2012
|
|
4DN4
 
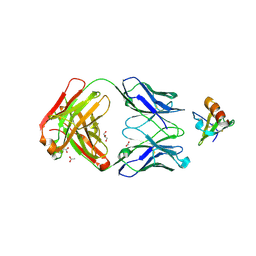 | | Crystal structure of the complex between cnto888 fab and mcp-1 mutant p8a | | Descriptor: | ACETATE ION, C-C motif chemokine 2, CNTO888 HEAVY CHAIN, ... | | Authors: | Obmolova, G, Teplyakov, A, Malia, T, Grygiel, T, Sweet, R, Snyder, L, Gilliland, G. | | Deposit date: | 2012-02-08 | | Release date: | 2012-10-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for high selectivity of anti-CCL2 neutralizing antibody CNTO 888.
Mol.Immunol., 51, 2012
|
|
4QHU
 
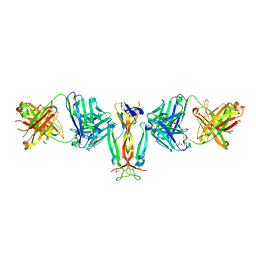 | | Crystal Structure of IL-17A/Fab6785 complex | | Descriptor: | CHLORIDE ION, Fab6785 heavy chain, Fab6785 light chain, ... | | Authors: | Luo, J, Gilliland, G.L, Malia, T, Obmolova, G, Teplyakov, A. | | Deposit date: | 2014-05-29 | | Release date: | 2015-08-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | IL-17A Asymmetry in a Complex with a Neutralizing Antibody Fab6785 Reveals a Potential Mechanism of Receptor Differentiation
To be Published
|
|
1B7D
 
 | | NEUROTOXIN (TS1) FROM BRAZILIAN SCORPION TITYUS SERRULATUS | | Descriptor: | PHOSPHATE ION, PROTEIN (NEUROTOXIN TS1) | | Authors: | Polikarpov, I, Sanches Jr, M.S, Marangoni, S, Toyama, M.H, Teplyakov, A. | | Deposit date: | 1999-01-21 | | Release date: | 1999-07-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of neurotoxin Ts1 from Tityus serrulatus provides insights into the specificity and toxicity of scorpion toxins.
J.Mol.Biol., 290, 1999
|
|
2HWG
 
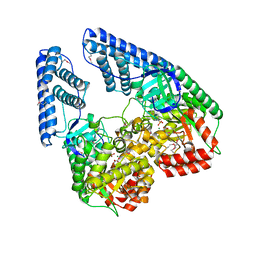 | | Structure of phosphorylated Enzyme I of the phosphoenolpyruvate:sugar phosphotransferase system | | Descriptor: | MAGNESIUM ION, OXALATE ION, Phosphoenolpyruvate-protein phosphotransferase | | Authors: | Lim, K, Teplyakov, A, Herzberg, O. | | Deposit date: | 2006-08-01 | | Release date: | 2006-11-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of phosphorylated enzyme I, the phosphoenolpyruvate:sugar phosphotransferase system sugar translocation signal protein.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1XFF
 
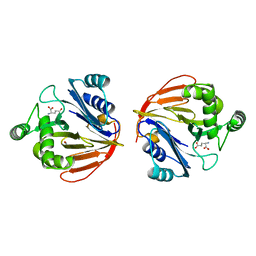 | | Glutaminase domain of glucosamine 6-phosphate synthase complexed with glutamate | | Descriptor: | ACETATE ION, GLUTAMIC ACID, Glucosamine--fructose-6-phosphate aminotransferase [isomerizing], ... | | Authors: | Isupov, M.N, Teplyakov, A. | | Deposit date: | 2004-09-14 | | Release date: | 2004-09-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Substrate Binding is Required for Assembly of the Active Conformation of the Catalytic Site in Ntn Amidotransferases: Evidence from the 1.8 Angstrom Crystal Structure of the Glutaminase Domain of Glucosamine 6-Phosphate Synthase
Structure, 4, 1996
|
|
1XFG
 
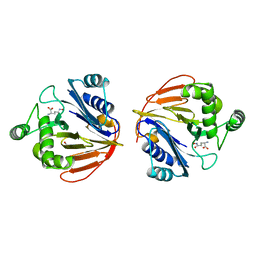 | | Glutaminase domain of glucosamine 6-phosphate synthase complexed with l-glu hydroxamate | | Descriptor: | ACETATE ION, GLUTAMINE HYDROXAMATE, Glucosamine--fructose-6-phosphate aminotransferase [isomerizing], ... | | Authors: | Isupov, M.N, Teplyakov, A. | | Deposit date: | 2004-09-14 | | Release date: | 2004-09-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate Binding is Required for Assembly of the Active Conformation of the Catalytic Site in Ntn Amidotransferases: Evidence from the 1.8 Angstrom Crystal Structure of the Glutaminase Domain of Glucosamine 6-Phosphate Synthase
Structure, 4, 1996
|
|
5E2V
 
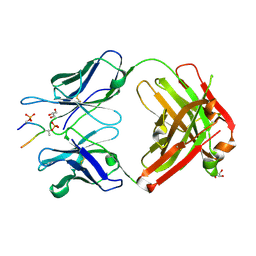 | |
5E2U
 
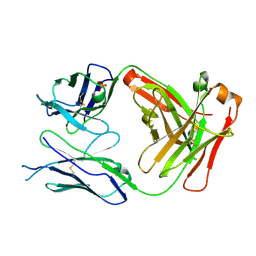 | |
