5M1H
 
 | |
4JNH
 
 | |
4JMR
 
 | | A unique spumavirus gag N-terminal domain with functional properties of orthoretroviral Matrix and Capsid | | Descriptor: | Env protein, Gag protein | | Authors: | Taylor, I.A, Goldstone, D.C, Flower, T.G, Ball, N.J. | | Deposit date: | 2013-03-14 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A Unique Spumavirus Gag N-terminal Domain with Functional Properties of Orthoretroviral Matrix and Capsid.
Plos Pathog., 9, 2013
|
|
1MB1
 
 | | MBP1 FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | MLU1-BOX BINDING PROTEIN | | Authors: | Taylor, I.A, Smerdon, S.J. | | Deposit date: | 1997-07-23 | | Release date: | 1998-07-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The X-ray structure of the DNA-binding domain from the Saccharomyces cerevisiae cell-cycle transcription factor Mbp1 at 2.1 A resolution.
J.Mol.Biol., 272, 1997
|
|
5M1G
 
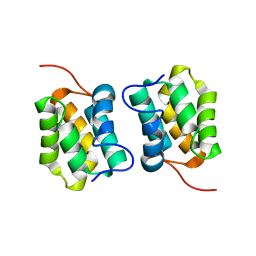 | |
7ZUD
 
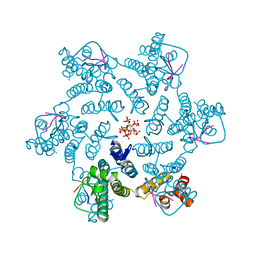 | | Crystal structure of HIV-1 capsid IP6-CPSF6 complex | | Descriptor: | Capsid protein p24, Cleavage and polyadenylation specificity factor subunit 6, INOSITOL HEXAKISPHOSPHATE | | Authors: | Nicastro, G, Taylor, I.A. | | Deposit date: | 2022-05-12 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | CP-MAS and Solution NMR Studies of Allosteric Communication in CA-assemblies of HIV-1.
J.Mol.Biol., 434, 2022
|
|
4UX5
 
 | | Structure of DNA complex of PCG2 | | Descriptor: | 5'-D(*CP*AP*AP*TP*GP*AP*CP*GP*CP*GP*TP*AP*AP*GP)-3', 5'-D(*CP*TP*TP*AP*CP*GP*CP*GP*TP*CP*AP*TP*TP*GP)-3', TRANSCRIPTION FACTOR MBP1 | | Authors: | Liu, J, Huang, J, Zhao, Y, Liu, H, Wang, D, Yang, J, Zhao, W, Taylor, I.A, Peng, Y. | | Deposit date: | 2014-08-19 | | Release date: | 2015-01-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of DNA Recognition by Pcg2 Reveals a Novel DNA Binding Mode for Winged Helix-Turn-Helix Domains.
Nucleic Acids Res., 43, 2015
|
|
7A5Y
 
 | | Crystal structure of tetrameric human H215A-SAMHD1 (residues 109-626) with Rp-dGTP-alphaS (T8T) and Mg | | Descriptor: | 2'-deoxyguanosine-5'-O-(1-thiotriphosphate), Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, FE (III) ION, ... | | Authors: | Morris, E.R, Kunzelmann, S, Caswell, S.J, Purkiss, A, Taylor, I.A. | | Deposit date: | 2020-08-24 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Probing the Catalytic Mechanism and Inhibition of SAMHD1 Using the Differential Properties of R p - and S p -dNTP alpha S Diastereomers.
Biochemistry, 60, 2021
|
|
6ZUE
 
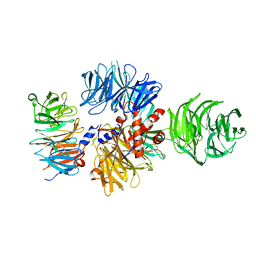 | |
2V4X
 
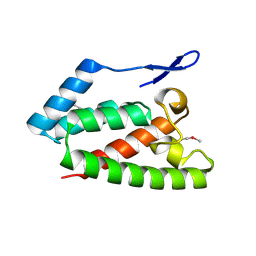 | | Crystal Structure of Jaagsiekte Sheep Retrovirus Capsid N-terminal domain | | Descriptor: | CAPSID PROTEIN P27 | | Authors: | Mortuza, G.B, Goldstone, D.C, Pashley, C, Haire, L.F, Palmarini, M, Taylor, W.R, Stoye, J.P, Taylor, I.A. | | Deposit date: | 2008-09-30 | | Release date: | 2008-11-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the Capsid Amino-Terminal Domain from the Betaretrovirus, Jaagsiekte Sheep Retrovirus.
J.Mol.Biol., 386, 2009
|
|
2XGV
 
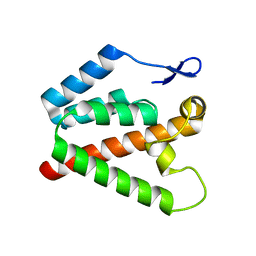 | | Structure of the N-terminal domain of capsid protein from Rabbit Endogenous Lentivirus (RELIK) | | Descriptor: | PSIV CAPSID N-TERMINAL DOMAIN | | Authors: | Goldstone, D.C, Robertson, L.E, Haire, L.F, Stoye, J.P, Taylor, I.A. | | Deposit date: | 2010-06-07 | | Release date: | 2010-09-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Analysis of Prehistoric Lentiviruses Uncovers an Ancient Molecular Interface.
Cell Host Microbe, 8, 2010
|
|
2XGU
 
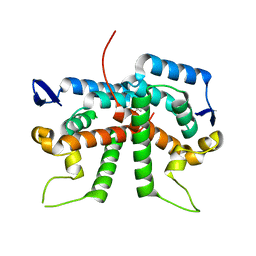 | | Structure of the N-terminal domain of capsid protein from Rabbit Endogenous Lentivirus (RELIK) | | Descriptor: | ACETATE ION, RELIK CAPSID N-TERMINAL DOMAIN | | Authors: | Goldstone, D.C, Taylor, I.A, Robertson, L.E, Haire, L.F, Stoye, J.P. | | Deposit date: | 2010-06-07 | | Release date: | 2010-09-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Structural and Functional Analysis of Prehistoric Lentiviruses Uncovers an Ancient Molecular Interface.
Cell Host Microbe, 8, 2010
|
|
2XGY
 
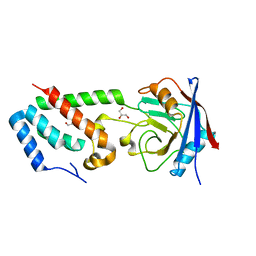 | | Complex of Rabbit Endogenous Lentivirus (RELIK)Capsid with Cyclophilin A | | Descriptor: | GLYCEROL, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A, RELIK CAPSID N-TERMINAL DOMAIN | | Authors: | Goldstone, D.C, Robertson, L.E, Haire, L.F, Stoye, J.P, Taylor, I.A. | | Deposit date: | 2010-06-08 | | Release date: | 2010-09-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Functional Analysis of Prehistoric Lentiviruses Uncovers an Ancient Molecular Interface.
Cell Host Microbe, 8, 2010
|
|
1L3G
 
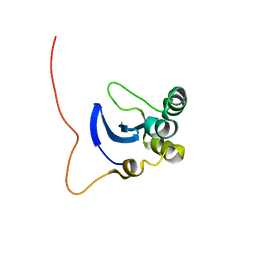 | | NMR Structure of the DNA-binding Domain of Cell Cycle Protein, Mbp1(2-124) from Saccharomyces cerevisiae | | Descriptor: | TRANSCRIPTION FACTOR Mbp1 | | Authors: | Nair, M, McIntosh, P.B, Frenkiel, T.A, Kelly, G, Taylor, I.A, Smerdon, S.J, Lane, A.N. | | Deposit date: | 2002-02-27 | | Release date: | 2003-02-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the DNA-Binding Domain of the Cell Cycle Protein Mbp1 from Saccharomyces cerevisiae
Biochemistry, 42, 2003
|
|
9HTS
 
 | |
5Z1V
 
 | | Crystal structure of AvrPib | | Descriptor: | AvrPib protein | | Authors: | Zhang, X, He, D, Zhao, Y.X, Taylor, I.A, Peng, Y.L, Yang, J, Liu, J.F. | | Deposit date: | 2017-12-28 | | Release date: | 2018-09-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.661 Å) | | Cite: | A positive-charged patch and stabilized hydrophobic core are essential for avirulence function of AvrPib in the rice blast fungus.
Plant J., 96, 2018
|
|
6TXC
 
 | | Crystal structure of tetrameric human wt-SAMHD1 (residues 109-626) with GTP, dATP, dCMPNPP and Mg | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | Authors: | Morris, E.R, Kunzelmann, S, Caswell, S.J, Arnold, L.H, Purkiss, A, Kelly, G, Taylor, I.A. | | Deposit date: | 2020-01-14 | | Release date: | 2020-06-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Crystal structures of SAMHD1 inhibitor complexes reveal the mechanism of water-mediated dNTP hydrolysis.
Nat Commun, 11, 2020
|
|
6TXF
 
 | | Crystal structure of tetrameric human D137N-SAMHD1 (residues 109-626) with XTP, dAMPNPP and Mn | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, FE (III) ION, ... | | Authors: | Morris, E.R, Kunzelmann, S, Caswell, S.J, Arnold, L.H, Purkiss, A.G, Kelly, G, Taylor, I.A. | | Deposit date: | 2020-01-14 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of SAMHD1 inhibitor complexes reveal the mechanism of water-mediated dNTP hydrolysis.
Nat Commun, 11, 2020
|
|
6TX0
 
 | | Crystal structure of tetrameric human D137N-SAMHD1 (residues 109-626) with XTP, dAMPNPP and Mg | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, FE (III) ION, ... | | Authors: | Morris, E.R, Kunzelmann, S, Caswell, S.J, Arnold, L.H, Purkiss, A.G, Kelly, G, Taylor, I.A. | | Deposit date: | 2020-01-13 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structures of SAMHD1 inhibitor complexes reveal the mechanism of water-mediated dNTP hydrolysis.
Nat Commun, 11, 2020
|
|
6ES4
 
 | | A cryptic RNA-binding domain mediates Syncrip recognition and exosomal partitioning of miRNA targets | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Syncrip, ... | | Authors: | Hobor, F, Dallmann, A, Ball, N.J, Cicchini, C, Battistelli, C, Ogrodowicz, R.W, Christodoulou, E, Martin, S.R, Castello, A, Tripodi, M, Taylor, I.A, Ramos, A. | | Deposit date: | 2017-10-19 | | Release date: | 2018-03-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A cryptic RNA-binding domain mediates Syncrip recognition and exosomal partitioning of miRNA targets.
Nat Commun, 9, 2018
|
|
4CC9
 
 | | Crystal structure of human SAMHD1 (amino acid residues 582-626) bound to Vpx isolated from sooty mangabey and human DCAF1 (amino acid residues 1058-1396) | | Descriptor: | DEOXYNUCLEOSIDE TRIPHOSPHATE TRIPHOSPHOHYDROLASE SAMHD1, PROTEIN VPRBP, PROTEIN VPX, ... | | Authors: | Schwefel, D, Groom, H.C.T, Boucherit, V.C, Christodoulou, E, Walker, P.A, Stoye, J.P, Bishop, K.N, Taylor, I.A. | | Deposit date: | 2013-10-19 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.473 Å) | | Cite: | Structural Basis of Lentiviral Subversion of a Cellular Protein Degradation Pathway.
Nature, 505, 2014
|
|
1G6G
 
 | |
7NLG
 
 | |
7NLI
 
 | |
7NLH
 
 | |
