8TZZ
 
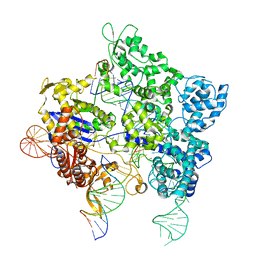 | | SpG Cas9 with NGC PAM DNA target | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(P*CP*GP*TP*TP*TP*GP*TP*AP*CP*TP*GP*CP*AP*GP*CP*G)-3'), DNA (5'-D(P*TP*CP*TP*CP*AP*TP*CP*TP*TP*TP*AP*TP*GP*CP*GP*TP*C)-3'), ... | | Authors: | Bravo, J.P.K, Hibshman, G.N, Taylor, D.W. | | Deposit date: | 2023-08-28 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Unraveling the mechanisms of PAMless DNA interrogation by SpRY-Cas9.
Nat Commun, 15, 2024
|
|
3J04
 
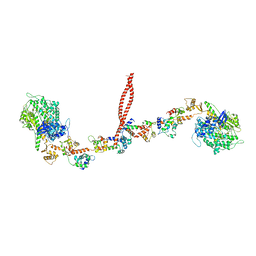 | | EM structure of the heavy meromyosin subfragment of Chick smooth muscle Myosin with regulatory light chain in phosphorylated state | | Descriptor: | Myosin light polypeptide 6, Myosin regulatory light chain 2, smooth muscle major isoform, ... | | Authors: | Baumann, B.A.J, Taylor, D, Huang, Z, Tama, F, Fagnant, P.M, Trybus, K, Taylor, K. | | Deposit date: | 2011-02-18 | | Release date: | 2011-11-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Phosphorylated smooth muscle heavy meromyosin shows an open conformation linked to activation.
J.Mol.Biol., 415, 2012
|
|
9BQV
 
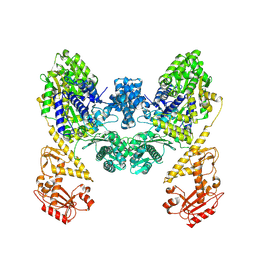 | | DdmD dimer apoprotein | | Descriptor: | Helicase/UvrB N-terminal domain-containing protein | | Authors: | Bravo, J.P.K, Taylor, D.W. | | Deposit date: | 2024-05-10 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Plasmid targeting and destruction by the DdmDE bacterial defence system.
Nature, 630, 2024
|
|
6NTS
 
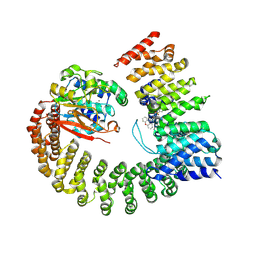 | | Protein Phosphatase 2A (Aalpha-B56alpha-Calpha) holoenzyme in complex with a Small Molecule Activator of PP2A (SMAP) | | Descriptor: | MANGANESE (II) ION, N-[(1R,2R,3S)-2-hydroxy-3-(10H-phenoxazin-10-yl)cyclohexyl]-4-(trifluoromethoxy)benzene-1-sulfonamide, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit alpha isoform, ... | | Authors: | Huang, W, Taylor, D. | | Deposit date: | 2019-01-30 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Selective PP2A Enhancement through Biased Heterotrimer Stabilization.
Cell, 181, 2020
|
|
2DFS
 
 | | 3-D structure of Myosin-V inhibited state | | Descriptor: | Calmodulin, Myosin-5A | | Authors: | Liu, J, Taylor, D.W, Krementsova, E.B, Trybus, K.M, Taylor, K.A. | | Deposit date: | 2006-03-03 | | Release date: | 2006-04-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (24 Å) | | Cite: | Three-dimensional structure of the myosin V inhibited state by cryoelectron tomography
Nature, 442, 2006
|
|
7UG6
 
 | | Cryo-EM structure of pre-60S ribosomal subunit, unmethylated G2922 | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Yelland, J.N, Bravo, J.P.K, Black, J.J.B, Taylor, D.W, Johnson, A.W. | | Deposit date: | 2022-03-24 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | A single 2'-O-methylation of ribosomal RNA gates assembly of a functional ribosome.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7UO0
 
 | | E.coli RNaseP Holoenzyme with Mg2+ | | Descriptor: | CALCIUM ION, Precursor tRNA substrate G(-1) G(-2), RNase P RNA, ... | | Authors: | Huang, W, Taylor, D.J. | | Deposit date: | 2022-04-12 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural and mechanistic basis for recognition of alternative tRNA precursor substrates by bacterial ribonuclease P.
Nat Commun, 13, 2022
|
|
7UO1
 
 | | E.coli RNaseP Holoenzyme with Mg2+ | | Descriptor: | CALCIUM ION, E.coli RNase P RNA, Precursor tRNA substrate U(-1) and A(-2), ... | | Authors: | Huang, W, Taylor, D.J. | | Deposit date: | 2022-04-12 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and mechanistic basis for recognition of alternative tRNA precursor substrates by bacterial ribonuclease P.
Nat Commun, 13, 2022
|
|
7UO2
 
 | | E.coli RNaseP Holoenzyme with Mg2+ | | Descriptor: | MAGNESIUM ION, RNase P RNA, Ribonuclease P protein component | | Authors: | Huang, W, Taylor, D.J. | | Deposit date: | 2022-04-12 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and mechanistic basis for recognition of alternative tRNA precursor substrates by bacterial ribonuclease P.
Nat Commun, 13, 2022
|
|
7UO5
 
 | | E.coli RNaseP Holoenzyme with Mg2+ | | Descriptor: | CALCIUM ION, RNase P RNA, Ribonuclease P protein component | | Authors: | Huang, W, Taylor, D.J. | | Deposit date: | 2022-04-12 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and mechanistic basis for recognition of alternative tRNA precursor substrates by bacterial ribonuclease P.
Nat Commun, 13, 2022
|
|
7TUF
 
 | | Crystal structure of Tapasin in complex with PaSta1-Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PaSta1 Fab heavy chain, PaSta1 Fab kappa light chain, ... | | Authors: | Jiang, J, Natarajan, K, Taylor, D.K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2022-02-02 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural mechanism of tapasin-mediated MHC-I peptide loading in antigen presentation.
Nat Commun, 13, 2022
|
|
7TUG
 
 | | Crystal structure of Tapasin in complex with PaSta2-Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PaSta2 Fab heavy chain, PaSta2 Fab kappa light chain, ... | | Authors: | Jiang, J, Natarajan, K, Taylor, D.K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2022-02-02 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural mechanism of tapasin-mediated MHC-I peptide loading in antigen presentation.
Nat Commun, 13, 2022
|
|
7TUH
 
 | | Crystal structure of anti-tapasin PaSta2-Fab | | Descriptor: | PaSta2 Fab heavy chain, PaSta2 Fab kappa light chain | | Authors: | Jiang, J, Natarajan, K, Taylor, D.K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2022-02-02 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural mechanism of tapasin-mediated MHC-I peptide loading in antigen presentation.
Nat Commun, 13, 2022
|
|
6MDR
 
 | | Cryo-EM structure of the Ceru+32/GFP-17 protomer | | Descriptor: | Ceru+32, GFP-17 | | Authors: | Simon, A.J, Zhou, Y, Ramasubramani, V, Glaser, J, Pothukuchy, A, Golihar, J, Gerberich, J.C, Leggere, J.C, Morrow, B.R, Jung, C, Glotzer, S.C, Taylor, D.W, Ellington, A.D. | | Deposit date: | 2018-09-05 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Supercharging enables organized assembly of synthetic biomolecules.
Nat Chem, 11, 2019
|
|
7KHA
 
 | | Cryo-EM Structure of the Desulfovibrio vulgaris Type I-C Apo Cascade | | Descriptor: | CRISPR-associated protein, CT1133 family, CT1134 family, ... | | Authors: | O'Brien, R, Wrapp, D, Bravo, J.P.K, Schwartz, E, Taylor, D. | | Deposit date: | 2020-10-20 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural basis for assembly of non-canonical small subunits into type I-C Cascade.
Nat Commun, 11, 2020
|
|
6N8K
 
 | | Cryo-EM structure of early cytoplasmic-immediate (ECI) pre-60S ribosomal subunit | | Descriptor: | 5.8S rRNA, 5S rRNA, 60S ribosomal export protein NMD3, ... | | Authors: | Zhou, Y, Musalgaonkar, S, Johnson, A.W, Taylor, D.W. | | Deposit date: | 2018-11-29 | | Release date: | 2019-03-13 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Tightly-orchestrated rearrangements govern catalytic center assembly of the ribosome.
Nat Commun, 10, 2019
|
|
6N8O
 
 | | Cryo-EM structure of Rpl10-inserted (RI) pre-60S ribosomal subunit | | Descriptor: | 5.8S rRNA, 5S rRNA, 60S ribosomal export protein NMD3, ... | | Authors: | Zhou, Y, Musalgaonkar, S, Johnson, A.W, Taylor, D.W. | | Deposit date: | 2018-11-29 | | Release date: | 2019-03-13 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Tightly-orchestrated rearrangements govern catalytic center assembly of the ribosome.
Nat Commun, 10, 2019
|
|
6N8J
 
 | | Cryo-EM structure of late nuclear (LN) pre-60S ribosomal subunit | | Descriptor: | 5.8S rRNA, 5S rRNA, 60S ribosomal protein L11-A, ... | | Authors: | Zhou, Y, Musalgaonkar, S, Johnson, A.W, Taylor, D.W. | | Deposit date: | 2018-11-29 | | Release date: | 2019-03-13 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Tightly-orchestrated rearrangements govern catalytic center assembly of the ribosome.
Nat Commun, 10, 2019
|
|
6N8L
 
 | | Cryo-EM structure of early cytoplasmic-late (ECL) pre-60S ribosomal subunit | | Descriptor: | 5.8S rRNA, 5S rRNA, 60S ribosomal export protein NMD3, ... | | Authors: | Zhou, Y, Musalgaonkar, S, Johnson, A.W, Taylor, D.W. | | Deposit date: | 2018-11-29 | | Release date: | 2019-03-13 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Tightly-orchestrated rearrangements govern catalytic center assembly of the ribosome.
Nat Commun, 10, 2019
|
|
6N8M
 
 | | Cryo-EM structure of pre-Lsg1 (PL) pre-60S ribosomal subunit | | Descriptor: | 5.8S RNA, 5S rRNA, 60S ribosomal export protein NMD3, ... | | Authors: | Zhou, Y, Musalgaonkar, S, Johnson, A.W, Taylor, D.W. | | Deposit date: | 2018-11-29 | | Release date: | 2019-03-13 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Tightly-orchestrated rearrangements govern catalytic center assembly of the ribosome.
Nat Commun, 10, 2019
|
|
6N8N
 
 | | Cryo-EM structure of Lsg1-engaged (LE) pre-60S ribosomal subunit | | Descriptor: | 5.8S rRNA, 5S rRNA, 60S ribosomal export protein NMD3, ... | | Authors: | Zhou, Y, Musalgaonkar, S, Johnson, A.W, Taylor, D.W. | | Deposit date: | 2018-11-29 | | Release date: | 2019-03-13 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Tightly-orchestrated rearrangements govern catalytic center assembly of the ribosome.
Nat Commun, 10, 2019
|
|
1SJJ
 
 | |
7MDW
 
 | |
1I84
 
 | | CRYO-EM STRUCTURE OF THE HEAVY MEROMYOSIN SUBFRAGMENT OF CHICKEN GIZZARD SMOOTH MUSCLE MYOSIN WITH REGULATORY LIGHT CHAIN IN THE DEPHOSPHORYLATED STATE. ONLY C ALPHAS PROVIDED FOR REGULATORY LIGHT CHAIN. ONLY BACKBONE ATOMS PROVIDED FOR S2 FRAGMENT. | | Descriptor: | SMOOTH MUSCLE MYOSIN ESSENTIAL LIGHT CHAIN, SMOOTH MUSCLE MYOSIN HEAVY CHAIN, SMOOTH MUSCLE MYOSIN REGULATORY LIGHT CHAIN | | Authors: | Wendt, T, Taylor, D, Trybus, K.M, Taylor, K. | | Deposit date: | 2001-03-12 | | Release date: | 2001-03-28 | | Last modified: | 2022-12-21 | | Method: | ELECTRON CRYSTALLOGRAPHY (20 Å) | | Cite: | Three-dimensional image reconstruction of dephosphorylated smooth muscle heavy meromyosin reveals asymmetry in the interaction between myosin heads and placement of subfragment 2.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
8SYF
 
 | | Homology model of Acto-HMM complex in ADP-state. Chicken smooth muscle HMM and chicken pectoralis actin | | Descriptor: | Actin, alpha skeletal muscle, Myosin light polypeptide 6, ... | | Authors: | Hojjatian, A, Taylor, D.W, Daneshparvar, N, Trybus, K.M, Taylor, K.A. | | Deposit date: | 2023-05-25 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (19 Å) | | Cite: | Double-headed binding of myosin II to F-actin shows the effect of strain on head structure.
J.Struct.Biol., 215, 2023
|
|
