4V7H
 
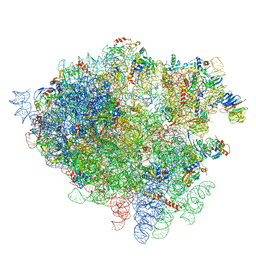 | | Structure of the 80S rRNA and proteins and P/E tRNA for eukaryotic ribosome based on cryo-EM map of Thermomyces lanuginosus ribosome at 8.9A resolution | | Descriptor: | 18S rRNA, 26S ribosomal RNA, 40S ribosomal protein S0(A), ... | | Authors: | Taylor, D.J, Devkota, B, Huang, A.D, Topf, M, Narayanan, E, Sali, A, Harvey, S.C, Frank, J. | | Deposit date: | 2009-09-22 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Comprehensive molecular structure of the eukaryotic ribosome.
Structure, 17, 2009
|
|
2P8X
 
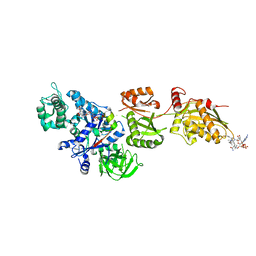 | | Fitted structure of ADPR-eEF2 in the 80S:ADPR-eEF2:GDPNP cryo-EM reconstruction | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Elongation factor 2, Elongation factor Tu-B, ... | | Authors: | Taylor, D.J, Nilsson, J, Merrill, A.R, Andersen, G.R, Nissen, P, Frank, J. | | Deposit date: | 2007-03-23 | | Release date: | 2007-05-08 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (9.7 Å) | | Cite: | Structures of modified eEF2.80S ribosome complexes reveal the role of GTP hydrolysis in translocation.
Embo J., 26, 2007
|
|
2P8Y
 
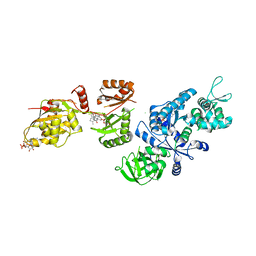 | | Fitted structure of ADPR-eEF2 in the 80S:ADPR-eEF2:GDP:sordarin cryo-EM reconstruction | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Elongation factor 2, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Taylor, D.J, Nilsson, J, Merrill, A.R, Andersen, G.R, Nissen, P, Frank, J. | | Deposit date: | 2007-03-23 | | Release date: | 2007-05-08 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (11.7 Å) | | Cite: | Structures of modified eEF2.80S ribosome complexes reveal the role of GTP hydrolysis in translocation.
Embo J., 26, 2007
|
|
7R6Z
 
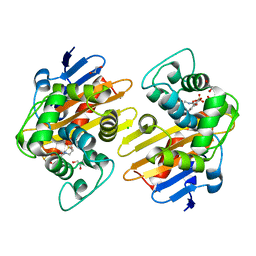 | | OXA-48 bound by Compound 3.3 | | Descriptor: | 1,2-ETHANEDIOL, 4-amino-5-hydroxynaphthalene-2,7-disulfonic acid, Beta-lactamase, ... | | Authors: | Taylor, D.M, Hu, L, Prasad, B.V.V, Sankaran, B, Palzkill, T. | | Deposit date: | 2021-06-24 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unique Diacidic Fragments Inhibit the OXA-48 Carbapenemase and Enhance the Killing of Escherichia coli Producing OXA-48.
Acs Infect Dis., 7, 2021
|
|
6XQR
 
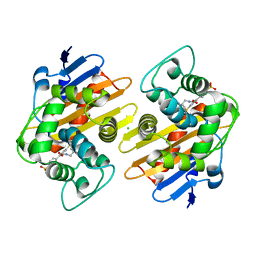 | | OXA-48 bound by Compound 2.2 | | Descriptor: | Beta-lactamase, CHLORIDE ION, [1,1'-biphenyl]-4,4'-disulfonic acid | | Authors: | Taylor, D.M, Hu, L, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2020-07-10 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unique Diacidic Fragments Inhibit the OXA-48 Carbapenemase and Enhance the Killing of Escherichia coli Producing OXA-48.
Acs Infect Dis., 7, 2021
|
|
2P8W
 
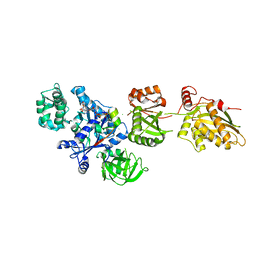 | | Fitted structure of eEF2 in the 80S:eEF2:GDPNP cryo-EM reconstruction | | Descriptor: | Elongation factor 2, Elongation factor Tu-B, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Taylor, D.J, Nilsson, J, Merrill, A.R, Andersen, G.R, Nissen, P, Frank, J. | | Deposit date: | 2007-03-23 | | Release date: | 2007-05-08 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (11.3 Å) | | Cite: | Structures of modified eEF2.80S ribosome complexes reveal the role of GTP hydrolysis in translocation.
Embo J., 26, 2007
|
|
6UVK
 
 | | OXA-48 bound by inhibitor CDD-97 | | Descriptor: | 1,2-ETHANEDIOL, 1-{4-[4-(2-ethoxyphenyl)piperazin-1-yl]-1,3,5-triazin-2-yl}piperidine-4-carboxylic acid, Beta-lactamase, ... | | Authors: | Taylor, D.M, Hu, L, Prasad, B.V.V, Sankaran, B, Palzkill, T.G. | | Deposit date: | 2019-11-02 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identifying Oxacillinase-48 Carbapenemase Inhibitors Using DNA-Encoded Chemical Libraries.
Acs Infect Dis., 6, 2020
|
|
4CLN
 
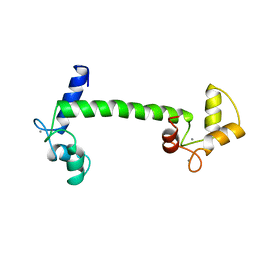 | | STRUCTURE OF A RECOMBINANT CALMODULIN FROM DROSOPHILA MELANOGASTER REFINED AT 2.2-ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, CALMODULIN | | Authors: | Taylor, D.A, Sack, J.S, Maune, J.F, Beckingham, K, Quiocho, F.A. | | Deposit date: | 1991-06-24 | | Release date: | 1992-07-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a recombinant calmodulin from Drosophila melanogaster refined at 2.2-A resolution.
J.Biol.Chem., 266, 1991
|
|
2P8Z
 
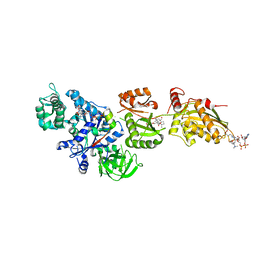 | | Fitted structure of ADPR-eEF2 in the 80S:ADPR-eEF2:GDPNP:sordarin cryo-EM reconstruction | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Elongation factor 2, Elongation factor Tu-B, ... | | Authors: | Taylor, D.J, Nilsson, J, Merrill, A.R, Andersen, G.R, Nissen, P, Frank, J. | | Deposit date: | 2007-03-23 | | Release date: | 2007-05-08 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Structures of modified eEF2.80S ribosome complexes reveal the role of GTP hydrolysis in translocation.
Embo J., 26, 2007
|
|
7K5V
 
 | | OXA-48 bound by Compound 3.1 | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Taylor, D.M, Hu, L, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2020-09-17 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Unique Diacidic Fragments Inhibit the OXA-48 Carbapenemase and Enhance the Killing of Escherichia coli Producing OXA-48.
Acs Infect Dis., 7, 2021
|
|
7JHQ
 
 | | OXA-48 bound by Compound 2.3 | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase OXA-48, CHLORIDE ION, ... | | Authors: | Taylor, D.M, Hu, L, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2020-07-21 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Unique Diacidic Fragments Inhibit the OXA-48 Carbapenemase and Enhance the Killing of Escherichia coli Producing OXA-48.
Acs Infect Dis., 7, 2021
|
|
7L8O
 
 | | OXA-48 bound by Compound 4.3 | | Descriptor: | 1,2-ETHANEDIOL, 9H-fluorene-2,7-disulfonate, Beta-lactamase, ... | | Authors: | Taylor, D.M, Hu, L, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2020-12-31 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Unique Diacidic Fragments Inhibit the OXA-48 Carbapenemase and Enhance the Killing of Escherichia coli Producing OXA-48.
Acs Infect Dis., 7, 2021
|
|
8S9X
 
 | |
8FZR
 
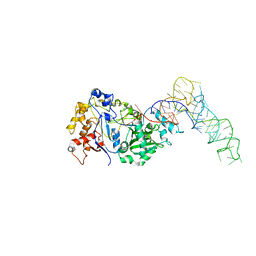 | |
8EW5
 
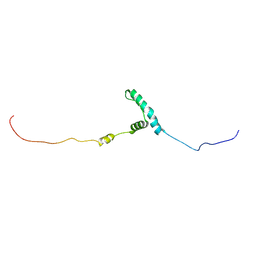 | | The structure of flightin within myosin thick filaments from Bombus ignitus flight muscle | | Descriptor: | Flightin | | Authors: | Li, J, Rahmani, H, Abbasi Yeganeh, F, Rastegarpouyani, H, Taylor, D.W, Wood, N.B, Previs, M.J, Iwamoto, H, Taylor, K.A. | | Deposit date: | 2022-10-21 | | Release date: | 2023-01-04 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structure of the Flight Muscle Thick Filament from the Bumble Bee, Bombus ignitus , at 6 angstrom Resolution.
Int J Mol Sci, 24, 2022
|
|
8W2S
 
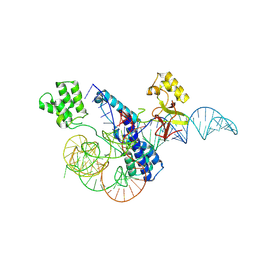 | |
3J04
 
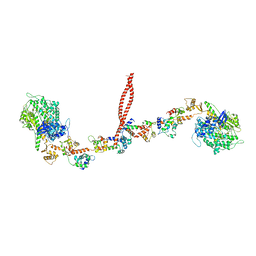 | | EM structure of the heavy meromyosin subfragment of Chick smooth muscle Myosin with regulatory light chain in phosphorylated state | | Descriptor: | Myosin light polypeptide 6, Myosin regulatory light chain 2, smooth muscle major isoform, ... | | Authors: | Baumann, B.A.J, Taylor, D, Huang, Z, Tama, F, Fagnant, P.M, Trybus, K, Taylor, K. | | Deposit date: | 2011-02-18 | | Release date: | 2011-11-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Phosphorylated smooth muscle heavy meromyosin shows an open conformation linked to activation.
J.Mol.Biol., 415, 2012
|
|
8W2Z
 
 | | Cas9d 6bp R-loop Seed Complex | | Descriptor: | DNA Non-Target Strand, DNA Target Strand, HNH nuclease domain-containing protein, ... | | Authors: | Fregoso Ocampo, R, Bravo, J.P.K, Taylor, D.W. | | Deposit date: | 2024-02-21 | | Release date: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | DNA targeting by compact Cas9d and its resurrected ancestor.
Nat Commun, 16, 2025
|
|
8SYF
 
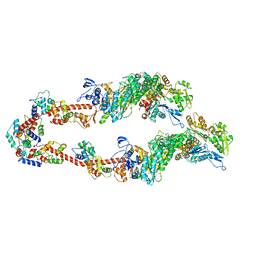 | | Homology model of Acto-HMM complex in ADP-state. Chicken smooth muscle HMM and chicken pectoralis actin | | Descriptor: | Actin, alpha skeletal muscle, Myosin light polypeptide 6, ... | | Authors: | Hojjatian, A, Taylor, D.W, Daneshparvar, N, Trybus, K.M, Taylor, K.A. | | Deposit date: | 2023-05-25 | | Release date: | 2023-08-30 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (19 Å) | | Cite: | Double-headed binding of myosin II to F-actin shows the effect of strain on head structure.
J.Struct.Biol., 215, 2023
|
|
1I84
 
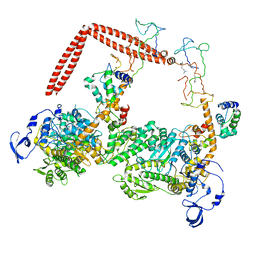 | | CRYO-EM STRUCTURE OF THE HEAVY MEROMYOSIN SUBFRAGMENT OF CHICKEN GIZZARD SMOOTH MUSCLE MYOSIN WITH REGULATORY LIGHT CHAIN IN THE DEPHOSPHORYLATED STATE. ONLY C ALPHAS PROVIDED FOR REGULATORY LIGHT CHAIN. ONLY BACKBONE ATOMS PROVIDED FOR S2 FRAGMENT. | | Descriptor: | SMOOTH MUSCLE MYOSIN ESSENTIAL LIGHT CHAIN, SMOOTH MUSCLE MYOSIN HEAVY CHAIN, SMOOTH MUSCLE MYOSIN REGULATORY LIGHT CHAIN | | Authors: | Wendt, T, Taylor, D, Trybus, K.M, Taylor, K. | | Deposit date: | 2001-03-12 | | Release date: | 2001-03-28 | | Last modified: | 2022-12-21 | | Method: | ELECTRON CRYSTALLOGRAPHY (20 Å) | | Cite: | Three-dimensional image reconstruction of dephosphorylated smooth muscle heavy meromyosin reveals asymmetry in the interaction between myosin heads and placement of subfragment 2.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
9BQV
 
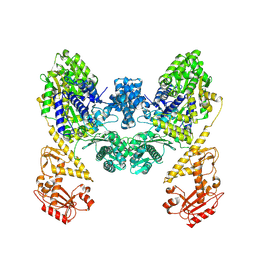 | | DdmD dimer apoprotein | | Descriptor: | Helicase/UvrB N-terminal domain-containing protein | | Authors: | Bravo, J.P.K, Taylor, D.W. | | Deposit date: | 2024-05-10 | | Release date: | 2024-07-03 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Plasmid targeting and destruction by the DdmDE bacterial defence system.
Nature, 630, 2024
|
|
9ARV
 
 | | CryoEM structure of AMETA-A3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin J chain, Isoform 1 of Immunoglobulin heavy constant mu | | Authors: | Huang, W, Sang, Z, Taylor, D. | | Deposit date: | 2024-02-23 | | Release date: | 2024-11-06 | | Last modified: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Adaptive multi-epitope targeting and avidity-enhanced nanobody platform for ultrapotent, durable antiviral therapy.
Cell, 187, 2024
|
|
1SJJ
 
 | |
7KOG
 
 | | Lethocerus Myosin II complete coiled-coil domain resolved in its native environment | | Descriptor: | Myosin heavy chain isoform Mhc_X1 | | Authors: | Rahmani, H, Hu, Z, Daneshparvar, N, Taylor, D, Taylor, K.A. | | Deposit date: | 2020-11-09 | | Release date: | 2021-03-24 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | The myosin II coiled-coil domain atomic structure in its native environment.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2DFS
 
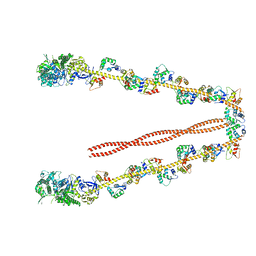 | | 3-D structure of Myosin-V inhibited state | | Descriptor: | Calmodulin, Myosin-5A | | Authors: | Liu, J, Taylor, D.W, Krementsova, E.B, Trybus, K.M, Taylor, K.A. | | Deposit date: | 2006-03-03 | | Release date: | 2006-04-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (24 Å) | | Cite: | Three-dimensional structure of the myosin V inhibited state by cryoelectron tomography
Nature, 442, 2006
|
|
