5KOW
 
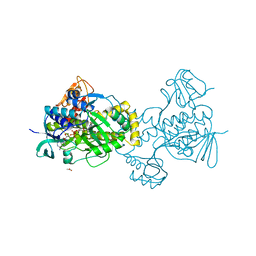 | | Structure of rifampicin monooxygenase | | 分子名称: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, Pentachlorophenol 4-monooxygenase | | 著者 | Tanner, J.J, Liu, L.-K. | | 登録日 | 2016-07-01 | | 公開日 | 2016-09-07 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The Structure of the Antibiotic Deactivating, N-hydroxylating Rifampicin Monooxygenase.
J.Biol.Chem., 291, 2016
|
|
5KOX
 
 | |
5HHF
 
 | |
6BSN
 
 | |
6D97
 
 | |
3HAZ
 
 | |
3ITG
 
 | |
2RBF
 
 | |
4ZVW
 
 | |
4ZVX
 
 | |
4ZVY
 
 | |
4O8A
 
 | |
6O4L
 
 | | Structure of ALDH7A1 mutant E399D complexed with NAD | | 分子名称: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde dehydrogenase, CHLORIDE ION, ... | | 著者 | Tanner, J.J, Korasick, D.A, Laciak, A.R. | | 登録日 | 2019-02-28 | | 公開日 | 2019-11-06 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural analysis of pathogenic mutations targeting Glu427 of ALDH7A1, the hot spot residue of pyridoxine-dependent epilepsy.
J. Inherit. Metab. Dis., 43, 2020
|
|
6O4D
 
 | | Structure of ALDH7A1 mutant W175A complexed with L-pipecolic acid | | 分子名称: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde dehydrogenase, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Tanner, J.J, Korasick, D.A, Laciak, A.R. | | 登録日 | 2019-02-28 | | 公開日 | 2019-07-24 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Structural and biochemical consequences of pyridoxine-dependent epilepsy mutations that target the aldehyde binding site of aldehyde dehydrogenase ALDH7A1.
Febs J., 287, 2020
|
|
6O4C
 
 | | Structure of ALDH7A1 mutant W175A complexed with NAD | | 分子名称: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde dehydrogenase, CHLORIDE ION, ... | | 著者 | Tanner, J.J, Korasick, D.A, Laciak, A.R. | | 登録日 | 2019-02-28 | | 公開日 | 2019-07-24 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural and biochemical consequences of pyridoxine-dependent epilepsy mutations that target the aldehyde binding site of aldehyde dehydrogenase ALDH7A1.
Febs J., 287, 2020
|
|
6O4K
 
 | | Structure of ALDH7A1 mutant E399Q complexed with NAD | | 分子名称: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde dehydrogenase, CHLORIDE ION, ... | | 著者 | Tanner, J.J, Korasick, D.A, Laciak, A.R. | | 登録日 | 2019-02-28 | | 公開日 | 2019-11-06 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Structural analysis of pathogenic mutations targeting Glu427 of ALDH7A1, the hot spot residue of pyridoxine-dependent epilepsy.
J. Inherit. Metab. Dis., 43, 2020
|
|
6O4I
 
 | | Structure of ALDH7A1 mutant E399D complexed with alpha-aminoadipate | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINOHEXANEDIOIC ACID, Alpha-aminoadipic semialdehyde dehydrogenase | | 著者 | Tanner, J.J, Korasick, D.A, Laciak, A.R. | | 登録日 | 2019-02-28 | | 公開日 | 2019-11-06 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural analysis of pathogenic mutations targeting Glu427 of ALDH7A1, the hot spot residue of pyridoxine-dependent epilepsy.
J. Inherit. Metab. Dis., 43, 2020
|
|
6O4B
 
 | | Structure of ALDH7A1 mutant W175G complexed with NAD | | 分子名称: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde dehydrogenase, CHLORIDE ION, ... | | 著者 | Tanner, J.J, Korasick, D.A, Laciak, A.R. | | 登録日 | 2019-02-28 | | 公開日 | 2019-07-24 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural and biochemical consequences of pyridoxine-dependent epilepsy mutations that target the aldehyde binding site of aldehyde dehydrogenase ALDH7A1.
Febs J., 287, 2020
|
|
1TJ2
 
 | | Crystal structure of E. coli PutA proline dehydrogenase domain (residues 86-669) complexed with acetate | | 分子名称: | ACETATE ION, Bifunctional putA protein, FLAVIN-ADENINE DINUCLEOTIDE | | 著者 | Tanner, J.J, Zhang, M, White, T.A, Schuermann, J.P, Baban, B.A, Becker, D.F. | | 登録日 | 2004-06-03 | | 公開日 | 2004-10-26 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structures of the Escherichia coli PutA proline dehydrogenase domain in complex with competitive inhibitors
Biochemistry, 43, 2004
|
|
1TJ1
 
 | | Crystal structure of E. coli PutA proline dehydrogenase domain (residues 86-669) complexed with L-lactate | | 分子名称: | (2S)-2-HYDROXYPROPANOIC ACID, Bifunctional putA protein, FLAVIN-ADENINE DINUCLEOTIDE | | 著者 | Tanner, J.J, Zhang, M, White, T.A, Schuermann, J.P, Baban, B.A, Becker, D.F. | | 登録日 | 2004-06-03 | | 公開日 | 2004-10-26 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structures of the Escherichia coli PutA proline dehydrogenase domain in complex with competitive inhibitors
Biochemistry, 43, 2004
|
|
3SME
 
 | | Structure of PTP1B inactivated by H2O2/bicarbonate | | 分子名称: | MAGNESIUM ION, Tyrosine-protein phosphatase non-receptor type 1 | | 著者 | Tanner, J.J, Singh, H. | | 登録日 | 2011-06-27 | | 公開日 | 2011-10-19 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The Biological Buffer Bicarbonate/CO(2) Potentiates H(2)O(2)-Mediated Inactivation of Protein Tyrosine Phosphatases.
J.Am.Chem.Soc., 133, 2011
|
|
1CER
 
 | |
8DKQ
 
 | |
8DKO
 
 | |
8DKP
 
 | |
