3Q35
 
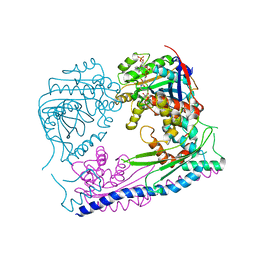 | | Structure of the Rtt109-AcCoA/Vps75 complex and implications for chaperone-mediated histone acetylation | | Descriptor: | 1,2-ETHANEDIOL, ACETYL COENZYME *A, Histone acetyltransferase, ... | | Authors: | Tang, Y, Yuan, H, Meeth, K, Marmorstein, R. | | Deposit date: | 2010-12-21 | | Release date: | 2011-02-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the Rtt109-AcCoA/Vps75 Complex and Implications for Chaperone-Mediated Histone Acetylation.
Structure, 19, 2011
|
|
3Q33
 
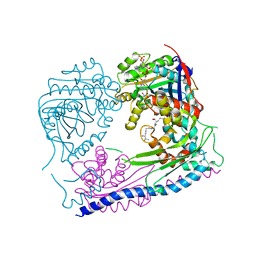 | | Structure of the Rtt109-AcCoA/Vps75 Complex and Implications for Chaperone-Mediated Histone Acetylation | | Descriptor: | 1,2-ETHANEDIOL, ACETYL COENZYME *A, HISTONE H3, ... | | Authors: | Tang, Y, Yuan, H, Meeth, K, Marmorstein, R. | | Deposit date: | 2010-12-21 | | Release date: | 2011-02-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Rtt109-AcCoA/Vps75 Complex and Implications for Chaperone-Mediated Histone Acetylation.
Structure, 19, 2011
|
|
5I1Q
 
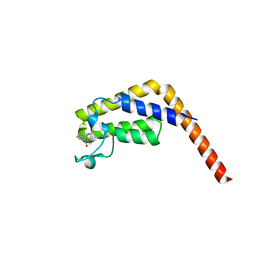 | | Second bromodomain of TAF1 bound to a pyrrolopyridone compound | | Descriptor: | 3-[6-(but-3-en-1-yl)-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-4-yl]-N,N-dimethylbenzamide, Transcription initiation factor TFIID subunit 1 | | Authors: | Tang, Y, Poy, F, Bellon, S.F. | | Deposit date: | 2016-01-09 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Diving into the Water: Inducible Binding Conformations for BRD4, TAF1(2), BRD9, and CECR2 Bromodomains.
J.Med.Chem., 59, 2016
|
|
5I29
 
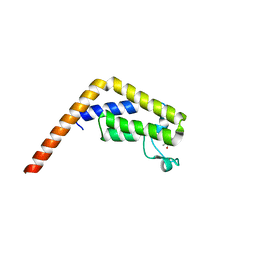 | | TAF1(2) bound to a pyrrolopyridone compound | | Descriptor: | CALCIUM ION, N,N-dimethyl-3-(6-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-4-yl)benzamide, Transcription initiation factor TFIID subunit 1 | | Authors: | Tang, Y, Poy, F, Bellon, S.F. | | Deposit date: | 2016-02-08 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Diving into the Water: Inducible Binding Conformations for BRD4, TAF1(2), BRD9, and CECR2 Bromodomains.
J.Med.Chem., 59, 2016
|
|
7TAB
 
 | | G-925 bound to the SMARCA4 (BRG1) Bromodomain | | Descriptor: | 2-(6-amino-5-phenylpyridazin-3-yl)phenol, Isoform 4 of Transcription activator BRG1 | | Authors: | Tang, Y, Poy, F, Taylor, A.M, Cochran, A.G, Bellon, S.F. | | Deposit date: | 2021-12-20 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | GNE-064: A Potent, Selective, and Orally Bioavailable Chemical Probe for the Bromodomains of SMARCA2 and SMARCA4 and the Fifth Bromodomain of PBRM1.
J.Med.Chem., 65, 2022
|
|
7TD9
 
 | | G-059 bound to the SMARCA4 (BRG1) Bromodomain | | Descriptor: | 4-phenyl-5H-pyridazino[4,3-b]indol-3-amine, Isoform 4 of Transcription activator BRG1 | | Authors: | Tang, Y, Poy, F, Taylor, A.M, Cochran, A.G, Bellon, S.F. | | Deposit date: | 2021-12-30 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | GNE-064: A Potent, Selective, and Orally Bioavailable Chemical Probe for the Bromodomains of SMARCA2 and SMARCA4 and the Fifth Bromodomain of PBRM1.
J.Med.Chem., 65, 2022
|
|
2HG4
 
 | | Structure of the ketosynthase-acyltransferase didomain of module 5 from DEBS. | | Descriptor: | 6-Deoxyerythronolide B Synthase, ACETATE ION, CHLORIDE ION, ... | | Authors: | Tang, Y, Kim, C.Y, Mathews, I.I, Cane, D.E, Khosla, C. | | Deposit date: | 2006-06-26 | | Release date: | 2006-07-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | The 2.7-A crystal structure of a 194-kDa homodimeric fragment of the 6-deoxyerythronolide B synthase.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3DM7
 
 | |
4RF2
 
 | |
4RF4
 
 | |
8T7Q
 
 | | Identification of GDC-1971 (RLY-1971), a SHP2 inhibitor designed for the treatment of solid tumors | | Descriptor: | 1-{3-[(2-chlorophenyl)sulfanyl]-1H-pyrazolo[3,4-b]pyrazin-6-yl}-4-methylpiperidin-4-amine, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Tang, Y, Nguyen, V, Wilbur, J.D. | | Deposit date: | 2023-06-21 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of GDC-1971 (RLY-1971), a SHP2 Inhibitor Designed for the Treatment of Solid Tumors.
J.Med.Chem., 66, 2023
|
|
8T8Q
 
 | | Identification of GDC-1971 (RLY-1971), a SHP2 inhibitor designed for the treatment of solid tumors | | Descriptor: | 1-[(3P)-3-(3-chloro-2-fluorophenyl)-1H-pyrazolo[3,4-b]pyrazin-6-yl]-4-methylpiperidin-4-amine, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Tang, Y, Nguyen, V, Wilbur, J.D. | | Deposit date: | 2023-06-23 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Identification of GDC-1971 (RLY-1971), a SHP2 Inhibitor Designed for the Treatment of Solid Tumors.
J.Med.Chem., 66, 2023
|
|
8T6D
 
 | | Identification of GDC-1971 (RLY-1971), a SHP2 inhibitor designed for the treatment of solid tumors | | Descriptor: | (3R)-1'-[3-(3,4-dihydro-1,5-naphthyridin-1(2H)-yl)-1H-pyrazolo[3,4-b]pyrazin-6-yl]-3H-spiro[[1]benzofuran-2,4'-piperidin]-3-amine, SULFATE ION, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Tang, Y, Nguyen, V, Wilbur, J.D. | | Deposit date: | 2023-06-15 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of GDC-1971 (RLY-1971), a SHP2 Inhibitor Designed for the Treatment of Solid Tumors.
J.Med.Chem., 66, 2023
|
|
8T6G
 
 | | Identification of GDC-1971 (RLY-1971), a SHP2 inhibitor designed for the treatment of solid tumors | | Descriptor: | (1S)-1-{6-[(1S)-1-amino-1,3-dihydrospiro[indene-2,4'-piperidin]-1'-yl]-3-(3,4-dihydro-1,5-naphthyridin-1(2H)-yl)-1H-pyrazolo[3,4-b]pyrazin-5-yl}ethan-1-ol, SULFATE ION, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Tang, Y, Nugyen, V, Wilbur, J.D. | | Deposit date: | 2023-06-15 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Identification of GDC-1971 (RLY-1971), a SHP2 Inhibitor Designed for the Treatment of Solid Tumors.
J.Med.Chem., 66, 2023
|
|
5KPE
 
 | | Solution NMR Structure of Denovo Beta Sheet Design Protein, Northeast Structural Genomics Consortium (NESG) Target OR664 | | Descriptor: | De novo Beta Sheet Design Protein OR664 | | Authors: | Tang, Y, Liu, G, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2016-07-03 | | Release date: | 2016-09-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
5KPH
 
 | |
1U5E
 
 | |
1U5D
 
 | | Crystal Structure of the PH domain of SKAP55 | | Descriptor: | SULFATE ION, Src Kinase-associated Phosphoprotein of 55 kDa | | Authors: | Tang, Y, Swanson, K.D, Neel, B.G, Eck, M.J. | | Deposit date: | 2004-07-27 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for the Dimerization and Phosphoinositide Specificity of the Src Kinase-associated Phosphoproteins SKAP55 and SKAP-Hom
To be Published
|
|
1U5G
 
 | |
1U5F
 
 | |
3R6L
 
 | | Caspase-2 T380A bound with Ac-VDVAD-CHO | | Descriptor: | Caspase-2 subunit p12, Caspase-2 subunit p18, Peptide Inhibitor (ACE)VDVAD-CHO | | Authors: | Tang, Y, Wells, J, Arkin, M. | | Deposit date: | 2011-03-21 | | Release date: | 2011-07-27 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and enzymatic insights into caspase-2 protein substrate recognition and catalysis.
J.Biol.Chem., 286, 2011
|
|
3R7B
 
 | | Caspase-2 bound to one copy of Ac-DVAD-CHO | | Descriptor: | Caspase-2 subunit p12, Caspase-2 subunit p18, Peptide Inhibitor (ACE)DVAD-CHO | | Authors: | Tang, Y, Wells, J, Arkin, M. | | Deposit date: | 2011-03-22 | | Release date: | 2011-07-27 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and enzymatic insights into caspase-2 protein substrate recognition and catalysis.
J.Biol.Chem., 286, 2011
|
|
3R6G
 
 | |
3R7S
 
 | | Crystal Structure of Apo Caspase2 | | Descriptor: | Caspase-2 subunit p12, Caspase-2 subunit p18 | | Authors: | Tang, Y, Wells, J, Arkin, M. | | Deposit date: | 2011-03-22 | | Release date: | 2011-07-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | Structural and enzymatic insights into caspase-2 protein substrate recognition and catalysis.
J.Biol.Chem., 286, 2011
|
|
3R7N
 
 | | Caspase-2 bound with two copies of Ac-DVAD-CHO | | Descriptor: | Caspase-2 subunit p12, Caspase-2 subunit p18, Peptide Inhibitor (ACE)DVAD-CHO | | Authors: | Tang, Y, Wells, J, Arkin, M. | | Deposit date: | 2011-03-22 | | Release date: | 2011-07-27 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural and enzymatic insights into caspase-2 protein substrate recognition and catalysis.
J.Biol.Chem., 286, 2011
|
|
